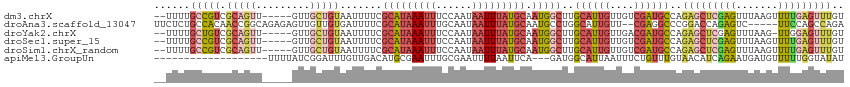
| Sequence ID | dm3.chrX |
|---|---|
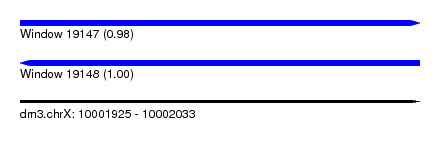
| Location | 10,001,925 – 10,002,033 |
| Length | 108 |
| Max. P | 0.998310 |

| Location | 10,001,925 – 10,002,033 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 68.02 |
| Shannon entropy | 0.61823 |
| G+C content | 0.38077 |
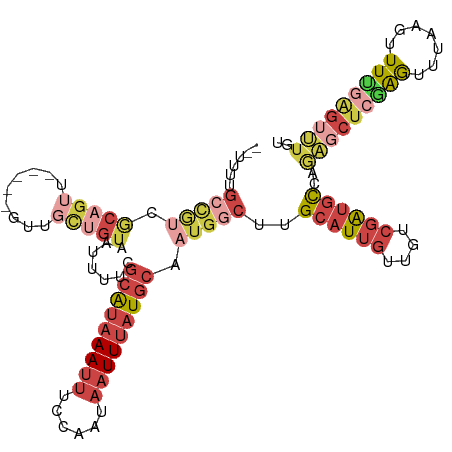
| Mean single sequence MFE | -23.10 |
| Consensus MFE | -9.25 |
| Energy contribution | -10.84 |
| Covariance contribution | 1.59 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.18 |
| SVM RNA-class probability | 0.984938 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
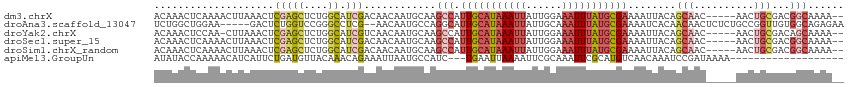
>dm3.chrX 10001925 108 + 22422827 ACAAACUCAAAACUUAAACUCGAGCUCUGGCAUCGACAACAAUGCAAGCCAUUGCAUAAAUUAUUGGAAAUUUAUGCGAAAAUUACAGCAAC-----AACUGCGACGGCAAAA-- ...................((((((....)).))))...........(((.(((((((((((......)))))))))))........(((..-----...)))...)))....-- ( -23.60, z-score = -2.91, R) >droAna3.scaffold_13047 835432 108 + 1816235 UCUGGCUGGAA-----GACUCUGGUCCGGGCCUCG--AACAAUGCCAGGCAUUGCAUAAAUUAUUGCAAAUUUAUGCGAAAAUCACAACAACUCUCUGCCGGUUGUGGCAGAGAA .(((((..((.-----...))..).))))((((.(--.......).)))).(((((((((((......))))))))))).............(((((((((....))))))))). ( -38.50, z-score = -3.34, R) >droYak2.chrX 18584071 107 + 21770863 ACAAACUCCAA-CUUAAACUCGAGCUCUGGCAUCGUCAACAAUGCAAGCCAUUGCAUAAAUUAUUGGAAAUUUAUGCGAAAAUUACAGCAAC-----AACUGCGACAGCAAAA-- ...........-...........(((.((((..(((.....)))...))))(((((((((((......))))))))))).......)))...-----...(((....)))...-- ( -19.40, z-score = -1.04, R) >droSec1.super_15 711023 108 + 1954846 ACAAACUCAAAACUUAAACUCGAGCUCUGGCAUCGACAACAAUGCAAGCCAUUGCAUAAAUUAUUGGAAAUUUAUGCGAAAAUUACAGCAAC-----AACUGCGACGGCAAAA-- ...................((((((....)).))))...........(((.(((((((((((......)))))))))))........(((..-----...)))...)))....-- ( -23.60, z-score = -2.91, R) >droSim1.chrX_random 2856684 108 + 5698898 ACAAACUCAAAACUUAAACUCGAGCUCUGGCAUCGACAACAAUGCAAGCCAUUGCAUAAAUUAUUGGAAAUUUAUGCGAAAAUUACAGCAAC-----AACUGCGACGGCAAAA-- ...................((((((....)).))))...........(((.(((((((((((......)))))))))))........(((..-----...)))...)))....-- ( -23.60, z-score = -2.91, R) >apiMel3.GroupUn 265387875 93 - 399230636 AUAUACCAAAAACAUCAUUCUGAUGUUACAAACAGAAAUUAAUGCCAUC---UGAAUUAAAAUUCGCAAAUUCGCAUGUCAACAAAUCCGAUAAAA------------------- ..........((((((.....))))))...............((.(((.---((((((..........)))))).))).))...............------------------- ( -9.90, z-score = -0.73, R) >consensus ACAAACUCAAAACUUAAACUCGAGCUCUGGCAUCGACAACAAUGCAAGCCAUUGCAUAAAUUAUUGGAAAUUUAUGCGAAAAUUACAGCAAC_____AACUGCGACGGCAAAA__ .......................(((.(.(((..........(((......(((((((((((......)))))))))))........)))..........))).).)))...... ( -9.25 = -10.84 + 1.59)
| Location | 10,001,925 – 10,002,033 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 68.02 |
| Shannon entropy | 0.61823 |
| G+C content | 0.38077 |
| Mean single sequence MFE | -30.60 |
| Consensus MFE | -13.99 |
| Energy contribution | -17.32 |
| Covariance contribution | 3.33 |
| Combinations/Pair | 1.41 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.32 |
| SVM RNA-class probability | 0.998310 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
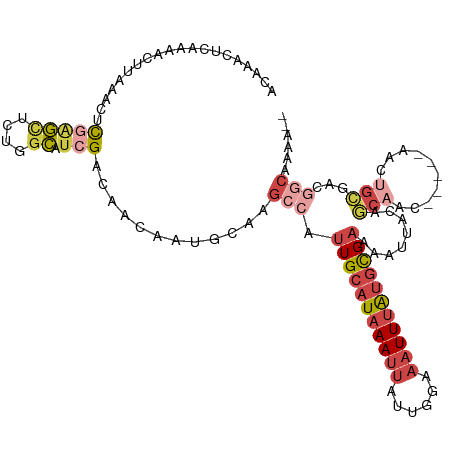
>dm3.chrX 10001925 108 - 22422827 --UUUUGCCGUCGCAGUU-----GUUGCUGUAAUUUUCGCAUAAAUUUCCAAUAAUUUAUGCAAUGGCUUGCAUUGUUGUCGAUGCCAGAGCUCGAGUUUAAGUUUUGAGUUUGU --....(((((.(((((.-----...))))).......(((((((((......))))))))).)))))..((((((....))))))..(((((((((.......))))))))).. ( -33.90, z-score = -3.83, R) >droAna3.scaffold_13047 835432 108 - 1816235 UUCUCUGCCACAACCGGCAGAGAGUUGUUGUGAUUUUCGCAUAAAUUUGCAAUAAUUUAUGCAAUGCCUGGCAUUGUU--CGAGGCCCGGACCAGAGUC-----UUCCAGCCAGA (((((((((......)))))))))..((((.((..((((((((((((......)))))))))....((.(((.((...--.)).))).))....)))..-----.)))))).... ( -36.30, z-score = -3.00, R) >droYak2.chrX 18584071 107 - 21770863 --UUUUGCUGUCGCAGUU-----GUUGCUGUAAUUUUCGCAUAAAUUUCCAAUAAUUUAUGCAAUGGCUUGCAUUGUUGACGAUGCCAGAGCUCGAGUUUAAG-UUGGAGUUUGU --....(((((.(((((.-----...))))).......(((((((((......))))))))).)))))..((((((....))))))..((((((.((......-)).)))))).. ( -29.50, z-score = -1.56, R) >droSec1.super_15 711023 108 - 1954846 --UUUUGCCGUCGCAGUU-----GUUGCUGUAAUUUUCGCAUAAAUUUCCAAUAAUUUAUGCAAUGGCUUGCAUUGUUGUCGAUGCCAGAGCUCGAGUUUAAGUUUUGAGUUUGU --....(((((.(((((.-----...))))).......(((((((((......))))))))).)))))..((((((....))))))..(((((((((.......))))))))).. ( -33.90, z-score = -3.83, R) >droSim1.chrX_random 2856684 108 - 5698898 --UUUUGCCGUCGCAGUU-----GUUGCUGUAAUUUUCGCAUAAAUUUCCAAUAAUUUAUGCAAUGGCUUGCAUUGUUGUCGAUGCCAGAGCUCGAGUUUAAGUUUUGAGUUUGU --....(((((.(((((.-----...))))).......(((((((((......))))))))).)))))..((((((....))))))..(((((((((.......))))))))).. ( -33.90, z-score = -3.83, R) >apiMel3.GroupUn 265387875 93 + 399230636 -------------------UUUUAUCGGAUUUGUUGACAUGCGAAUUUGCGAAUUUUAAUUCA---GAUGGCAUUAAUUUCUGUUUGUAACAUCAGAAUGAUGUUUUUGGUAUAU -------------------......(((((((((......)))))))))...........(((---((.(((((((..(((((..........)))))))))))))))))..... ( -16.10, z-score = 0.07, R) >consensus __UUUUGCCGUCGCAGUU_____GUUGCUGUAAUUUUCGCAUAAAUUUCCAAUAAUUUAUGCAAUGGCUUGCAUUGUUGUCGAUGCCAGAGCUCGAGUUUAAGUUUUGAGUUUGU ......(((((.(((((.........))))).......(((((((((......))))))))).)))))..((((((....))))))..(((((((((.......))))))))).. (-13.99 = -17.32 + 3.33)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:30:15 2011