| Sequence ID | dm3.chr2L |
|---|---|
| Location | 10,801,738 – 10,801,834 |
| Length | 96 |
| Max. P | 0.963865 |

| Location | 10,801,738 – 10,801,832 |
|---|---|
| Length | 94 |
| Sequences | 9 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 55.18 |
| Shannon entropy | 0.90130 |
| G+C content | 0.41356 |
| Mean single sequence MFE | -19.51 |
| Consensus MFE | -6.96 |
| Energy contribution | -7.31 |
| Covariance contribution | 0.35 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.963865 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
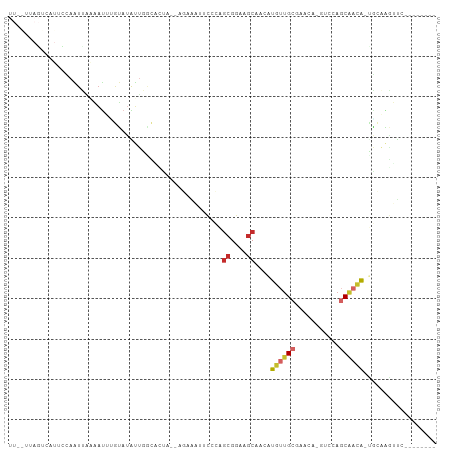
>dm3.chr2L 10801738 94 - 23011544 UUCGUUAGUCAUUCCAAUUAAAAUUUGUAUAUUGGCACUA--AGAAAUUCCCAGCGGAAGCAACAUGUUGCGAACA-GUCCAGCAACA-CGCAAGUUC-------- (((.(((((....(((((............))))).))))--)))).......((....)).....(((((.....-.....)))))(-(....))..-------- ( -18.50, z-score = -1.30, R) >droSim1.chr2L 10607767 94 - 22036055 UUAGUUAGUCAUUCCAAUUAAAAUUGGUAUAUUGGCACUA--AGAAAUUCCCAGCGGAAGCAACAUGUUGCGAAGA-GGCCAGCAACA-UGCAAGUUC-------- (((((..((((..(((((....))))).....))))))))--)..........((....))..((((((((.....-.....))))))-)).......-------- ( -24.10, z-score = -2.39, R) >droSec1.super_3 6219959 94 - 7220098 UUCGUUAGUCAUUUCAAUUAAAAUUUGUAUAUUGGCACUA--AGAAAUUCCCAGCGGAAGCAACAUGUUGCGAAAA-GCCCAGCAACA-UGCAAGUUC-------- .......((((...(((.......))).....))))(((.--...........((....))..((((((((.....-.....))))))-))..)))..-------- ( -19.10, z-score = -1.29, R) >droYak2.chr2L 7207909 94 - 22324452 UUCAUUAGUCAUUCCAAUUUAAAUUUGUAUAUUGGCACUA--AGAAAUUCCCAGCGGAAGCAACAUGUUGCGCACA-AUCCAGCAACA-CGCAAGUUC-------- (((.(((((....(((((.((......)).))))).))))--)))).......((....)).....(((((.....-.....)))))(-(....))..-------- ( -19.10, z-score = -2.00, R) >droEre2.scaffold_4929 12012495 94 + 26641161 UUCAUUAGUCAUUCCAAUCAAAAUUCGUAUAUUCGCCAUA--AGAAAUUCCCAGCGGAAGCAACAUGUUGCGAACA-GUCCAGCAACA-CGCAAGUUC-------- ..................................((....--...........((....))....((((((.....-.....))))))-.))......-------- ( -13.50, z-score = -1.05, R) >droAna3.scaffold_12916 14652069 74 - 16180835 ---------UUAAUAAGUUAUCAUAAAGAUGUUGCUUCGC---------UCCAGCGGUAGCAACAUGUUGCGAAUA-AAUCAGCAACAAUUUA------------- ---------....((((((.........((((((((((((---------....)))).))))))))(((((.....-.....)))))))))))------------- ( -22.20, z-score = -4.24, R) >droPer1.super_10 2020832 86 - 3432795 ------------UCGAAUAAACAUCGCUAAGCAGAUCUUGGGAGAGUGGUCGGACGUAGCCGGCGCA-AGCACGUGGGGCCAACUGUGGCUAUGAGCCG------- ------------((((....((.((.(((((.....)))))))..))..)))).(((((((.((...-.))..(..(......)..)))))))).....------- ( -24.60, z-score = 0.74, R) >droWil1.scaffold_180703 354132 92 + 3946847 ------------UUCAUUUGUUUGUUAGACAACAAUUUUU--UUUGUUGCCAAGCGGAAGCAACAUGUUGCCAAUAUUUAUAUCUCAACUCAUCGCUCUAAACGUU ------------....((((((((...(.((((((.....--.))))))))))))))).((((....))))................................... ( -13.50, z-score = -0.19, R) >droMoj3.scaffold_6500 13528555 84 + 32352404 -----AAAAAAUGUAAAUUG-AACGAAUAUGUUGCGGCCA---------CAGAGCGGAAGCAACAUGUUGCCAGCGCAGCCAGCAACAUUGCCAAUUCU------- -----...............-...((((((((((((((.(---------((..((....))....))).))).((...))..))))))).....)))).------- ( -21.00, z-score = -0.59, R) >consensus UU__UUAGUCAUUCCAAUUAAAAUUUGUAUAUUGGCACUA__AGAAAUUCCCAGCGGAAGCAACAUGUUGCGAACA_GUCCAGCAACA_UGCAAGUUC________ .....................................................((....))....((((((...........)))))).................. ( -6.96 = -7.31 + 0.35)

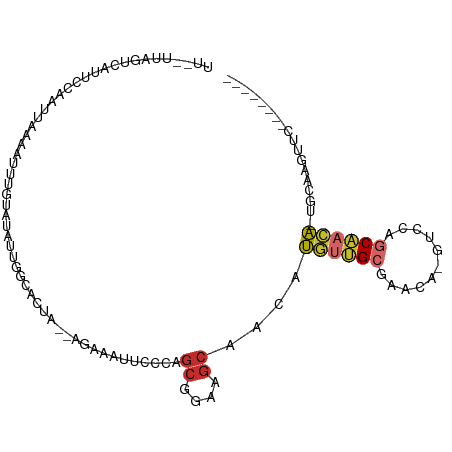
| Location | 10,801,738 – 10,801,834 |
|---|---|
| Length | 96 |
| Sequences | 9 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 55.23 |
| Shannon entropy | 0.89677 |
| G+C content | 0.40711 |
| Mean single sequence MFE | -19.96 |
| Consensus MFE | -6.96 |
| Energy contribution | -7.31 |
| Covariance contribution | 0.35 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.963067 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
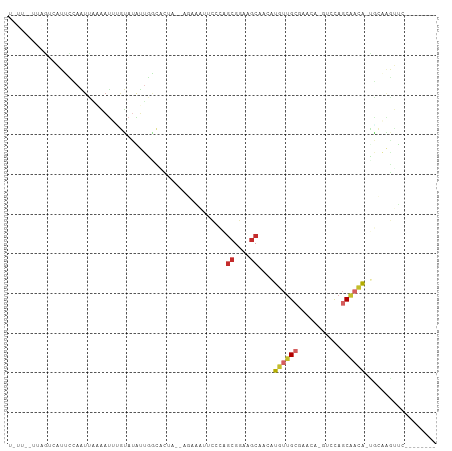
>dm3.chr2L 10801738 96 - 23011544 UUUUCGUUAGUCAUUCCAAUUAAAAUUUGUAUAUUGGCACUA--AGAAAUUCCCAGCGGAAGCAACAUGUUGCGAACA-GUCCAGCAACA-CGCAAGUUC-------- .((((.(((((....(((((............))))).))))--)))))......((....)).....(((((.....-.....)))))(-(....))..-------- ( -19.40, z-score = -1.59, R) >droSim1.chr2L 10607767 96 - 22036055 UAUUAGUUAGUCAUUCCAAUUAAAAUUGGUAUAUUGGCACUA--AGAAAUUCCCAGCGGAAGCAACAUGUUGCGAAGA-GGCCAGCAACA-UGCAAGUUC-------- ..(((((..((((..(((((....))))).....))))))))--)..........((....))..((((((((.....-.....))))))-)).......-------- ( -24.40, z-score = -2.42, R) >droSec1.super_3 6219959 96 - 7220098 UUUUCGUUAGUCAUUUCAAUUAAAAUUUGUAUAUUGGCACUA--AGAAAUUCCCAGCGGAAGCAACAUGUUGCGAAAA-GCCCAGCAACA-UGCAAGUUC-------- .((((....((((...(((.......))).....))))....--.))))......((....))..((((((((.....-.....))))))-)).......-------- ( -19.80, z-score = -1.41, R) >droYak2.chr2L 7207909 96 - 22324452 UUUUCAUUAGUCAUUCCAAUUUAAAUUUGUAUAUUGGCACUA--AGAAAUUCCCAGCGGAAGCAACAUGUUGCGCACA-AUCCAGCAACA-CGCAAGUUC-------- .((((.(((((....(((((.((......)).))))).))))--)))))......((....)).....(((((.....-.....)))))(-(....))..-------- ( -20.00, z-score = -2.32, R) >droEre2.scaffold_4929 12012495 96 + 26641161 UUUUCAUUAGUCAUUCCAAUCAAAAUUCGUAUAUUCGCCAUA--AGAAAUUCCCAGCGGAAGCAACAUGUUGCGAACA-GUCCAGCAACA-CGCAAGUUC-------- ....................................((....--...........((....))....((((((.....-.....))))))-.))......-------- ( -13.50, z-score = -0.92, R) >droAna3.scaffold_12916 14652069 76 - 16180835 ---------GAUUAAUAAGUUAUCAUAAAGAUGUUGCUUCGC---------UCCAGCGGUAGCAACAUGUUGCGAAUA-AAUCAGCAACAAUUUA------------- ---------......((((((.........((((((((((((---------....)))).))))))))(((((.....-.....)))))))))))------------- ( -22.20, z-score = -3.91, R) >droPer1.super_10 2020832 88 - 3432795 ------------UUUCGAAUAAACAUCGCUAAGCAGAUCUUGGGAGAGUGGUCGGACGUAGCCGGCGCA-AGCACGUGGGGCCAACUGUGGCUAUGAGCCG------- ------------.(((((....((.((.(((((.....)))))))..))..)))))(((((((.((...-.))..(..(......)..)))))))).....------- ( -25.80, z-score = 0.42, R) >droWil1.scaffold_180703 354132 94 + 3946847 ------------UUUUCAUUUGUUUGUUAGACAACAAUUUUU--UUUGUUGCCAAGCGGAAGCAACAUGUUGCCAAUAUUUAUAUCUCAACUCAUCGCUCUAAACGUU ------------......((((((((...(.((((((.....--.))))))))))))))).((((....))))................................... ( -13.50, z-score = -0.17, R) >droMoj3.scaffold_6500 13528555 84 + 32352404 -------AAAAAAUGUAAAUUG-AACGAAUAUGUUGCGGCCA---------CAGAGCGGAAGCAACAUGUUGCCAGCGCAGCCAGCAACAUUGCCAAUUCU------- -------...............-...((((((((((((((.(---------((..((....))....))).))).((...))..))))))).....)))).------- ( -21.00, z-score = -0.59, R) >consensus U_UU__UUAGUCAUUCCAAUUAAAAUUUGUAUAUUGGCACUA__AGAAAUUCCCAGCGGAAGCAACAUGUUGCGAACA_GUCCAGCAACA_UGCAAGUUC________ .......................................................((....))....((((((...........)))))).................. ( -6.96 = -7.31 + 0.35)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:31:02 2011