
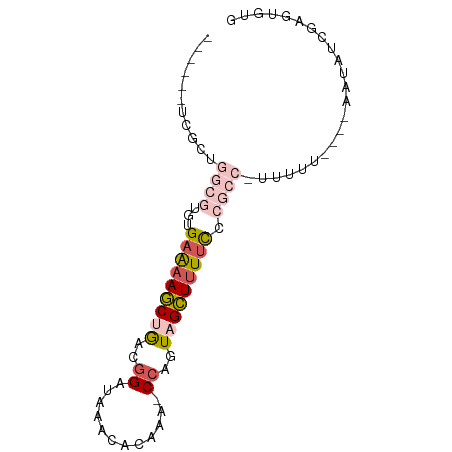
| Sequence ID | dm3.chrX |
|---|---|
| Location | 9,985,311 – 9,985,389 |
| Length | 78 |
| Max. P | 0.656775 |

| Location | 9,985,311 – 9,985,389 |
|---|---|
| Length | 78 |
| Sequences | 5 |
| Columns | 84 |
| Reading direction | forward |
| Mean pairwise identity | 66.19 |
| Shannon entropy | 0.55136 |
| G+C content | 0.46069 |
| Mean single sequence MFE | -18.40 |
| Consensus MFE | -7.56 |
| Energy contribution | -9.36 |
| Covariance contribution | 1.80 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.601761 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 9985311 78 + 22422827 -----UUCGCUGGCGUGUGAAAAGCUGACGGAUAAACACAAA-CCAGUAGCUUUUCCCGCCUUUUUUUCCCAAUAUCGAGUGUG -----..((((((((.(.(((((((((..((...........-))..)))))))))))))).........(......))))).. ( -19.90, z-score = -1.90, R) >droEre2.scaffold_4690 13626205 62 - 18748788 ------UCGCUGGCGUGUGAAAAGCUGACGGAUAAACACACA-CCAGUAGCUUUUC---------------AAUAUCGAGUGUG ------.((((.(....((((((((((..((...........-))..)))))))))---------------)....).)))).. ( -18.30, z-score = -1.96, R) >droYak2.chrX 18575052 72 + 21770863 ------GCGCUGGCGUGUGAAAAGCCGGCGGAUAAACACAAA-CCAGUAGCUUUUCCCGCCGUUUUUC---AAUAUCGAGUG-- ------.((((.(....((((((((.(((((..((((((...-...)).).)))..))))))))))))---)....).))))-- ( -22.40, z-score = -1.80, R) >droSec1.super_15 701571 75 + 1954846 ------UCGCUGGCGUGUGAAAAGCUGACGGAUAAACACAAA-CCAGUAGCUUUUCCCGCCUUUUUUUC--AAUAUCGAAUGUG ------.(((.((((.(.(((((((((..((...........-))..)))))))))))))).....(((--......))).))) ( -20.00, z-score = -2.27, R) >droWil1.scaffold_181130 904321 69 - 16660200 UGAGGCUUAAAGACCUAUGAGCAACUAGUAGAUCAACCCUCGUCCAGUUGUUUUCUU---------------GCAACAAGUUUG .((((.((...((.((((.((...)).)))).)))).)))).....(((((......---------------)))))....... ( -11.40, z-score = 0.17, R) >consensus ______UCGCUGGCGUGUGAAAAGCUGACGGAUAAACACAAA_CCAGUAGCUUUUCCCGCC_UUUUU____AAUAUCGAGUGUG ...........((((...(((((((((..((............))..))))))))).))))....................... ( -7.56 = -9.36 + 1.80)


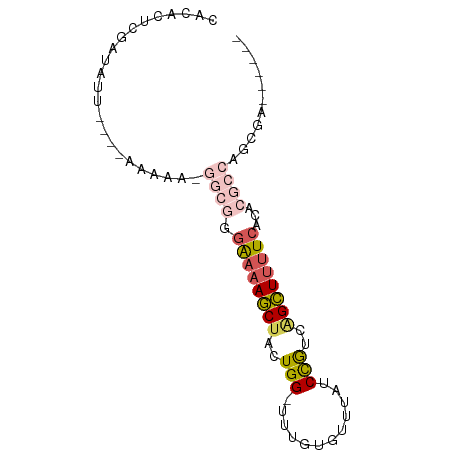
| Location | 9,985,311 – 9,985,389 |
|---|---|
| Length | 78 |
| Sequences | 5 |
| Columns | 84 |
| Reading direction | reverse |
| Mean pairwise identity | 66.19 |
| Shannon entropy | 0.55136 |
| G+C content | 0.46069 |
| Mean single sequence MFE | -18.65 |
| Consensus MFE | -9.22 |
| Energy contribution | -10.70 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.656775 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 9985311 78 - 22422827 CACACUCGAUAUUGGGAAAAAAAGGCGGGAAAAGCUACUGG-UUUGUGUUUAUCCGUCAGCUUUUCACACGCCAGCGAA----- ....((((....)))).......((((.((((((((((.((-(........))).)).))))))))...))))......----- ( -21.90, z-score = -1.68, R) >droEre2.scaffold_4690 13626205 62 + 18748788 CACACUCGAUAUU---------------GAAAAGCUACUGG-UGUGUGUUUAUCCGUCAGCUUUUCACACGCCAGCGA------ .............---------------.........((((-((((((.................))))))))))...------ ( -15.33, z-score = -1.20, R) >droYak2.chrX 18575052 72 - 21770863 --CACUCGAUAUU---GAAAAACGGCGGGAAAAGCUACUGG-UUUGUGUUUAUCCGCCGGCUUUUCACACGCCAGCGC------ --...........---.......((((.((((((((..(((-...........)))..))))))))...)))).....------ ( -20.40, z-score = -0.97, R) >droSec1.super_15 701571 75 - 1954846 CACAUUCGAUAUU--GAAAAAAAGGCGGGAAAAGCUACUGG-UUUGUGUUUAUCCGUCAGCUUUUCACACGCCAGCGA------ .............--........((((.((((((((((.((-(........))).)).))))))))...)))).....------ ( -20.60, z-score = -1.85, R) >droWil1.scaffold_181130 904321 69 + 16660200 CAAACUUGUUGC---------------AAGAAAACAACUGGACGAGGGUUGAUCUACUAGUUGCUCAUAGGUCUUUAAGCCUCA ....((.((((.---------------.......)))).))..((((.((((....(((........)))....)))).)))). ( -15.00, z-score = -0.41, R) >consensus CACACUCGAUAUU____AAAAA_GGCGGGAAAAGCUACUGG_UUUGUGUUUAUCCGUCAGCUUUUCACACGCCAGCGA______ .......................((((.((((((((..(((............)))..))))))))...))))........... ( -9.22 = -10.70 + 1.48)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:30:12 2011