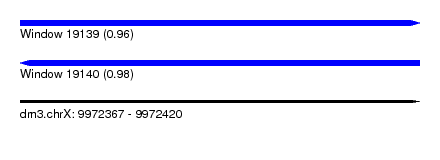
| Sequence ID | dm3.chrX |
|---|---|
| Location | 9,972,367 – 9,972,420 |
| Length | 53 |
| Max. P | 0.978578 |

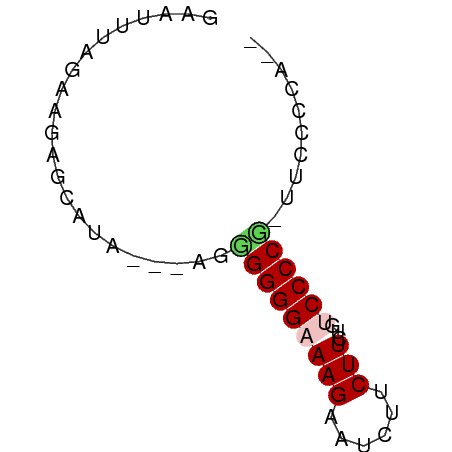
| Location | 9,972,367 – 9,972,420 |
|---|---|
| Length | 53 |
| Sequences | 5 |
| Columns | 59 |
| Reading direction | forward |
| Mean pairwise identity | 75.49 |
| Shannon entropy | 0.42780 |
| G+C content | 0.48121 |
| Mean single sequence MFE | -19.04 |
| Consensus MFE | -9.04 |
| Energy contribution | -9.76 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.960033 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 9972367 53 + 22422827 GAAUUUGGCAGAGCAUAGUGGGAGGGGAAAGAAUCAUCUU---UCCCCG-UUCCACA-- .................(((((((((((((((....))))---))))).-)))))).-- ( -23.90, z-score = -4.37, R) >droSim1.chrX 7939758 54 + 17042790 GAAUUCAGGAGAGCAUA--GAGUGGGGAAAGAAUCUUCUUCUUUCCCCG-UUCCCCA-- .......((.((((...--....((((((((((.....)))))))))))-))).)).-- ( -20.01, z-score = -2.42, R) >droSec1.super_15 688294 54 + 1954846 GAAUUUAGGAGAGCAUA--AAGUGGGGAAAGAAUCUUCUUCUUUCCCCG-UUCCCCA-- .......((.((((...--....((((((((((.....)))))))))))-))).)).-- ( -20.01, z-score = -2.93, R) >droYak2.chrX 18561873 56 + 21770863 GAUUUUAGAAAAGCACA---AAGGGGGCAAGAAUCUCCUUUCGCCCCCCCUUUCCCACC ................(---(((((((...(((......)))...))))))))...... ( -15.10, z-score = -1.58, R) >droEre2.scaffold_4690 13613353 53 - 18748788 GAUUUUAGAAGAGCAUA---AUGGGGGCAAGAAUCUACUUCUGCCCCCU-UUCCCCA-- .................---..(((((((.(((.....)))))))))).-.......-- ( -16.20, z-score = -1.73, R) >consensus GAAUUUAGAAGAGCAUA___AGGGGGGAAAGAAUCUUCUUCUGUCCCCG_UUCCCCA__ ......................((((((.((((.....)))).)))))).......... ( -9.04 = -9.76 + 0.72)

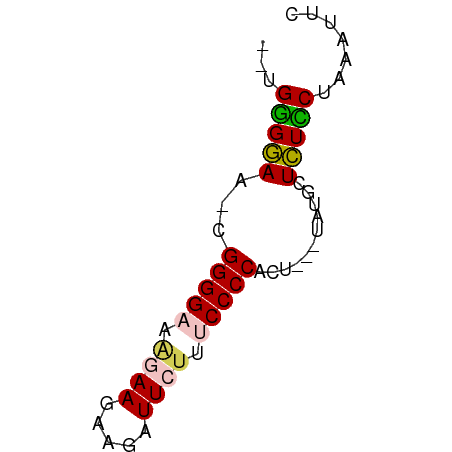
| Location | 9,972,367 – 9,972,420 |
|---|---|
| Length | 53 |
| Sequences | 5 |
| Columns | 59 |
| Reading direction | reverse |
| Mean pairwise identity | 75.49 |
| Shannon entropy | 0.42780 |
| G+C content | 0.48121 |
| Mean single sequence MFE | -20.34 |
| Consensus MFE | -10.42 |
| Energy contribution | -10.50 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.00 |
| SVM RNA-class probability | 0.978578 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 9972367 53 - 22422827 --UGUGGAA-CGGGGA---AAGAUGAUUCUUUCCCCUCCCACUAUGCUCUGCCAAAUUC --.((((..-.(((((---((((....)))))))))..))))................. ( -20.60, z-score = -4.27, R) >droSim1.chrX 7939758 54 - 17042790 --UGGGGAA-CGGGGAAAGAAGAAGAUUCUUUCCCCACUC--UAUGCUCUCCUGAAUUC --.(((((.-.((((((((((.....)))))))))).(..--...).)))))....... ( -20.60, z-score = -2.44, R) >droSec1.super_15 688294 54 - 1954846 --UGGGGAA-CGGGGAAAGAAGAAGAUUCUUUCCCCACUU--UAUGCUCUCCUAAAUUC --.(((((.-.((((((((((.....)))))))))).(..--...).)))))....... ( -20.60, z-score = -2.89, R) >droYak2.chrX 18561873 56 - 21770863 GGUGGGAAAGGGGGGGCGAAAGGAGAUUCUUGCCCCCUU---UGUGCUUUUCUAAAAUC ..(((((((((((((((((.((....)).))))))))).---....))))))))..... ( -21.40, z-score = -1.78, R) >droEre2.scaffold_4690 13613353 53 + 18748788 --UGGGGAA-AGGGGGCAGAAGUAGAUUCUUGCCCCCAU---UAUGCUCUUCUAAAAUC --.((((..-.((((((((((.....))).)))))))..---....))))......... ( -18.50, z-score = -1.73, R) >consensus __UGGGGAA_CGGGGAAAGAAGAAGAUUCUUUCCCCACU___UAUGCUCUCCUAAAUUC ...(((((...(((((.((((.....)))).)))))...........)))))....... (-10.42 = -10.50 + 0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:30:09 2011