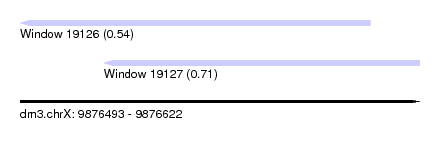
| Sequence ID | dm3.chrX |
|---|---|
| Location | 9,876,493 – 9,876,622 |
| Length | 129 |
| Max. P | 0.707016 |

| Location | 9,876,493 – 9,876,606 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 96.10 |
| Shannon entropy | 0.06966 |
| G+C content | 0.32029 |
| Mean single sequence MFE | -24.38 |
| Consensus MFE | -20.66 |
| Energy contribution | -20.74 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.539753 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
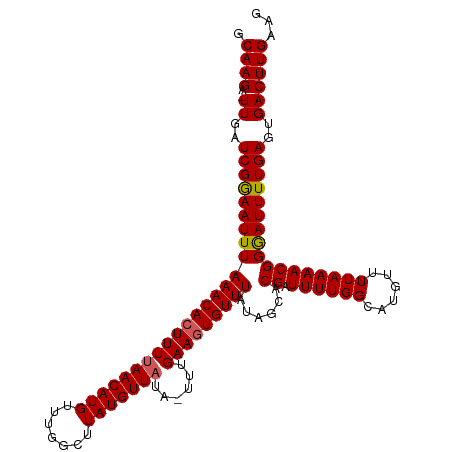
>dm3.chrX 9876493 113 - 22422827 GCAAGAUUUGAUCGGAAUUUAAACAGUUUUAACAUGUUUGGCUUAUGUUAUA-UUUGAAGUGUUUAUAGCACGAUUUUGGCAUGUUUUAAAACGGAAUUUUGAGUGACUUGAAG .((((.((...(((((((((.....(((((((((((((..(.(..(((((((-...........)))))))..).)..)))))))..)))))).)))))))))..))))))... ( -25.50, z-score = -2.00, R) >droSim1.chrX 7866142 112 - 17042790 GCAAGA-UUGAUCGGAAUUUAAACACUUUUAACAUGUUUGGCUUAUGUUAUA-UUUGAAGUGUUUAUAGCACGAUUUUGGCAUGUUUUAAAACGGGAUUUUGAGUGACUUGAAG .((((.-((..(((((((((((((((((((((((((.......)))))))..-...)))))))))).....((.((((((......)))))))).))))))))..))))))... ( -23.20, z-score = -1.42, R) >droSec1.super_130 25864 112 - 56455 GCAAGA-UUGAUCGGAAUUUAAACACUUUUAACAUGUUUGGCUUAUGUUAUA-UUUGAAGUGUUUAUAGCACGAUUUUGGCAUGUUUUAAAACGGGAUUUUGAGUGACUUGAAG .((((.-((..(((((((((((((((((((((((((.......)))))))..-...)))))))))).....((.((((((......)))))))).))))))))..))))))... ( -23.20, z-score = -1.42, R) >droYak2.chrX 18470944 112 - 21770863 GCAAGA-UUGAUCGGAAUUUAAACACUUUUAACAUGUUUGGCUUAUGUUUUA-UUUGAAGUGUUUAUAGCACGAUUUUGGCAUGUUUUAAAACGGGAUUUUGAGUGACUUGAAG .((((.-((..((((((((((((((((((.((((((.......))))))...-...)))))))))).....((.((((((......)))))))).))))))))..))))))... ( -22.20, z-score = -1.02, R) >droEre2.scaffold_4690 13516204 113 + 18748788 GCAAGA-UUGAUCGAAAUUUAAACACUUUUAACAUGGCUUGGCUAUGUUAUAUUUUGAAGUGUUUAUAGCACGAUUUUGGCAUGUUUUAAAACGGGAUUUUGAGUGACUUGAAG .((((.-((..(((((((((((((((((((((((((((...)))))))))......)))))))))).....((.((((((......)))))))).))))))))..))))))... ( -27.80, z-score = -2.87, R) >consensus GCAAGA_UUGAUCGGAAUUUAAACACUUUUAACAUGUUUGGCUUAUGUUAUA_UUUGAAGUGUUUAUAGCACGAUUUUGGCAUGUUUUAAAACGGGAUUUUGAGUGACUUGAAG .((((......(((((((((((((((((((((((((.......)))))))......)))))))))......((.((((((......)))))))))))))))))....))))... (-20.66 = -20.74 + 0.08)


| Location | 9,876,520 – 9,876,622 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 96.07 |
| Shannon entropy | 0.07009 |
| G+C content | 0.30769 |
| Mean single sequence MFE | -22.32 |
| Consensus MFE | -20.06 |
| Energy contribution | -20.46 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.707016 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
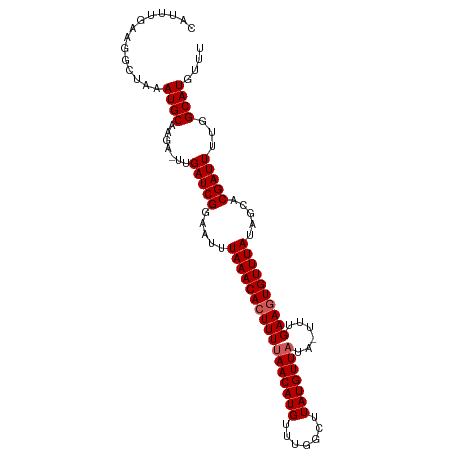
>dm3.chrX 9876520 102 - 22422827 CAUUUGAAGGCUAAAUGCAAGAUUUGAUCGGAAUUUAAACAGUUUUAACAUGUUUGGCUUAUGUUAUA-UUUGAAGUGUUUAUAGCACGAUUUUGGCAUGUUU ..............((((.......(((((.....((((((.((((((((((.......)))))))..-...))).)))))).....)))))...)))).... ( -18.30, z-score = -0.07, R) >droSim1.chrX 7866169 101 - 17042790 CAUUUGAAGGCUAAAUGCAAGA-UUGAUCGGAAUUUAAACACUUUUAACAUGUUUGGCUUAUGUUAUA-UUUGAAGUGUUUAUAGCACGAUUUUGGCAUGUUU ..............((((....-..(((((.....(((((((((((((((((.......)))))))..-...)))))))))).....)))))...)))).... ( -22.80, z-score = -1.91, R) >droSec1.super_130 25891 101 - 56455 CAUUUGAAGGCUAAAUGCAAGA-UUGAUCGGAAUUUAAACACUUUUAACAUGUUUGGCUUAUGUUAUA-UUUGAAGUGUUUAUAGCACGAUUUUGGCAUGUUU ..............((((....-..(((((.....(((((((((((((((((.......)))))))..-...)))))))))).....)))))...)))).... ( -22.80, z-score = -1.91, R) >droYak2.chrX 18470971 101 - 21770863 CAUUUGAAGGCUAAAUGCAAGA-UUGAUCGGAAUUUAAACACUUUUAACAUGUUUGGCUUAUGUUUUA-UUUGAAGUGUUUAUAGCACGAUUUUGGCAUGUUU ..............((((....-..(((((.....((((((((((.((((((.......))))))...-...)))))))))).....)))))...)))).... ( -21.80, z-score = -1.55, R) >droEre2.scaffold_4690 13516231 102 + 18748788 CAUUUGAAGGCUAAAUGCAAGA-UUGAUCGAAAUUUAAACACUUUUAACAUGGCUUGGCUAUGUUAUAUUUUGAAGUGUUUAUAGCACGAUUUUGGCAUGUUU ..............((((....-..(((((.....(((((((((((((((((((...)))))))))......)))))))))).....)))))...)))).... ( -25.90, z-score = -2.82, R) >consensus CAUUUGAAGGCUAAAUGCAAGA_UUGAUCGGAAUUUAAACACUUUUAACAUGUUUGGCUUAUGUUAUA_UUUGAAGUGUUUAUAGCACGAUUUUGGCAUGUUU ..............((((.......(((((.....(((((((((((((((((.......)))))))......)))))))))).....)))))...)))).... (-20.06 = -20.46 + 0.40)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:29:58 2011