| Sequence ID | dm3.chrX |
|---|---|
| Location | 9,859,446 – 9,859,510 |
| Length | 64 |
| Max. P | 0.661460 |

| Location | 9,859,446 – 9,859,510 |
|---|---|
| Length | 64 |
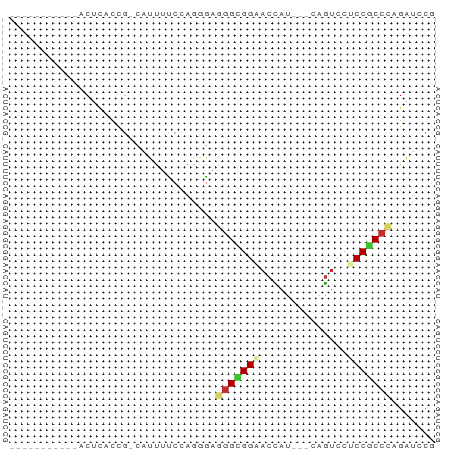
| Sequences | 9 |
| Columns | 68 |
| Reading direction | reverse |
| Mean pairwise identity | 73.11 |
| Shannon entropy | 0.48382 |
| G+C content | 0.59717 |
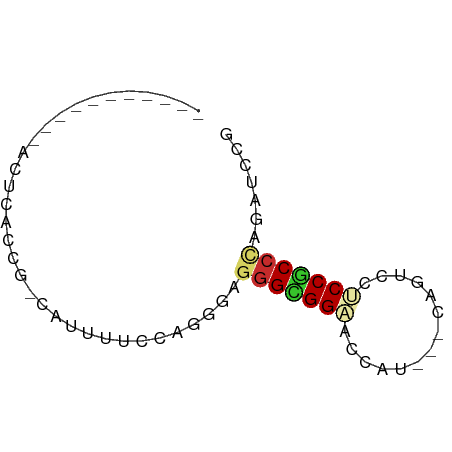
| Mean single sequence MFE | -13.17 |
| Consensus MFE | -9.02 |
| Energy contribution | -9.33 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.57 |
| Mean z-score | -0.86 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.661460 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chrX 9859446 64 - 22422827 GGUUUUUCCCAACUCACCG-CAUUUCCCAAGGAGGGCGGAACCAA---CAGUCCUCCGCCCAGAUCCG ((......)).........-..........((((((((((.....---......)))))))...))). ( -16.50, z-score = -1.23, R) >droSim1.chrX_random 2827507 53 - 5698898 -----------ACUCACCG-CAUUGCCCAAGGAGGGCGGAACCAU---CAGUCCUCCGCCCAGAUCCG -----------........-..........((((((((((.....---......)))))))...))). ( -15.90, z-score = -1.52, R) >droSec1.super_130 3020 53 - 56455 -----------ACUCACCG-CAUUGCCCAAGGAGGGCGGAACCAU---CAGUCCUCCGCCCAGAUCCG -----------........-..........((((((((((.....---......)))))))...))). ( -15.90, z-score = -1.52, R) >droYak2.chrX 18453141 53 - 21770863 -----------ACUGACCG-CAUUUUCCAGGGAGGGCGGAACCAU---CAGUCCUCCGCCCAGAUCCG -----------.(((....-.......)))((((((((((.....---......)))))))...))). ( -16.60, z-score = -1.12, R) >droEre2.scaffold_4690 13497893 53 + 18748788 -----------ACUUACCG-CAUUUUCCAAGGAGGGCGGAACCAU---CAGUCCUCCGCCUAGAUCCG -----------......((-.((((.....(((((((.((....)---).)))))))....)))).)) ( -14.60, z-score = -1.63, R) >droAna3.scaffold_13047 236616 53 - 1816235 -----------ACCUACCA-CGUUUUCCAGCGAAGGAGGCACUAC---CAGGCCGCCACCCAGGUUCA -----------((((....-(((......)))..((.(((.....---...))).))....))))... ( -12.60, z-score = -0.73, R) >dp4.chrXL_group3a 1089804 56 - 2690836 -----------ACUCACCG-AAUUAUCCAGAAUGGGUGGCACCAUCCCCAGUCCCCCGCCCAGAUCCG -----------........-.........((.(((((((.((........))...)))))))..)).. ( -11.00, z-score = -0.55, R) >droPer1.super_15 378362 57 + 2181545 -----------ACUCACCGAAAUUAUCCAGAAUGAGUGGCACCAUCCCCAGUCCCCCGCCCAGAUCCG -----------......................((.(((........))).))............... ( -4.10, z-score = 0.81, R) >droWil1.scaffold_181096 9385954 52 + 12416693 ------------CUUACUG-CAUCGUCGAGUGUCGGUGGGACUAU---UAAUCCACCACCUAGAUCCG ------------.......-....(((.((.((.((((((.....---...)))))))))).)))... ( -11.30, z-score = -0.28, R) >consensus ___________ACUCACCG_CAUUUUCCAGGGAGGGCGGAACCAU___CAGUCCUCCGCCCAGAUCCG .................................(((((((..............)))))))....... ( -9.02 = -9.33 + 0.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:29:55 2011