| Sequence ID | dm3.chr2L |
|---|---|
| Location | 947,004 – 947,104 |
| Length | 100 |
| Max. P | 0.971450 |

| Location | 947,004 – 947,104 |
|---|---|
| Length | 100 |
| Sequences | 11 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 76.68 |
| Shannon entropy | 0.50957 |
| G+C content | 0.45738 |
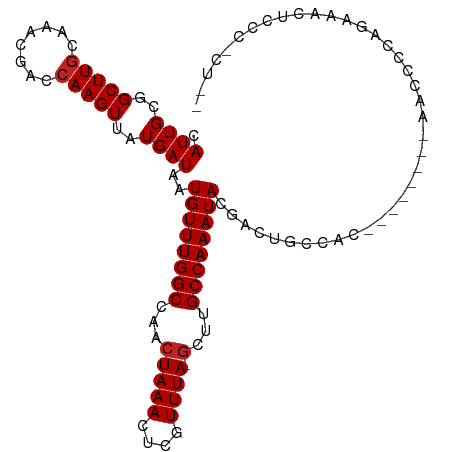
| Mean single sequence MFE | -21.55 |
| Consensus MFE | -14.65 |
| Energy contribution | -14.75 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.54 |
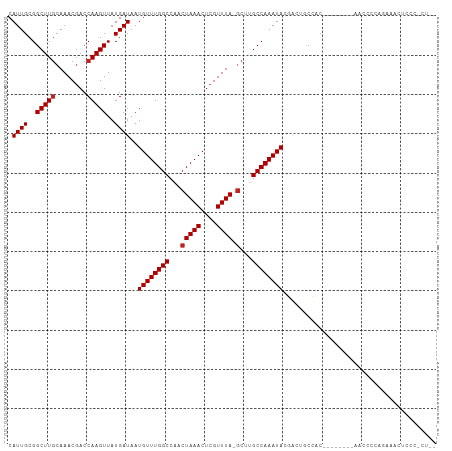
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.971450 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
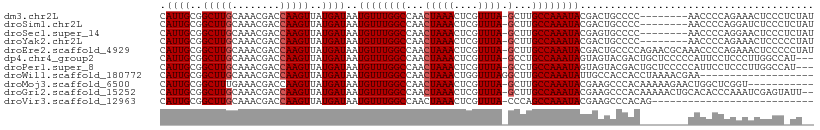
>dm3.chr2L 947004 100 - 23011544 CAUUGCGGCUUGCAAACGACCAAGUUAUGAUAAUGUUUGGCCAACUAAACUCGUUUA-GCUUGCCAAAUACGACUGCCCC--------AACCCCAGAAACUCCCUCUAU ..(((.(((...(.(((......))).......((((((((...(((((....))))-)...)))))))).)...))).)--------))................... ( -19.50, z-score = -1.79, R) >droSim1.chr2L 937805 100 - 22036055 CAUUGCGGCUUGCAAACGACCAAGUUAUGAUAAUGUUUGGCCAACUAAACUCGUUUA-GCUUGCCAAAUACGACUGCCCC--------AACCCCAGGAUCUCCCUCUAU ..(((.(((...(.(((......))).......((((((((...(((((....))))-)...)))))))).)...))).)--------))....(((.....))).... ( -20.70, z-score = -1.40, R) >droSec1.super_14 918037 100 - 2068291 CAUUGCGGCUUGCAAACGACCAAGUUAUGAUAAUGUUUGGCCAACUAAACUCGUUUA-GCUUGCCAAAUACGAGUGCCCC--------AACCCCAGGAACUCCCUCUAU .((((..(((((........)))))..))))..((((((((...(((((....))))-)...)))))))).((((..((.--------.......)).))))....... ( -23.10, z-score = -1.82, R) >droYak2.chr2L 934761 100 - 22324452 CAUUGCGGCUUGCAAACGACCAAGUUAUGAUAAUGUUUGGCCAACUAAACUCGUUUA-GCUUGCCAAAUACGACUGCCCC--------AACCCCAGAAACUCCCCCUAU ..(((.(((...(.(((......))).......((((((((...(((((....))))-)...)))))))).)...))).)--------))................... ( -19.50, z-score = -1.93, R) >droEre2.scaffold_4929 1008148 108 - 26641161 CAUUGCGGCUUGCAAACGACCAAGUUAUGAUAAUGUUUGGCCAACUAAACUCGUUUA-GCUUGCCAAAUACGACUGCCCCAGAACGCAAACCCCAGAAACUCCCCCUAU ..((((((((((........)))))........((((((((...(((((....))))-)...))))))))..............))))).................... ( -21.70, z-score = -1.83, R) >dp4.chr4_group2 846125 105 + 1235136 CAUUGCGGCUUGCAAACGACCAAGUUAUGAUAAUGUUUGGCCAACUAAACUCGUUUA-GCCUGCCAAAUAGUAGUACGACUGCUCCCCCAUUCCUCCCUUGGCCAU--- ..((((.....))))..(.(((((..(.((...((((((((...(((((....))))-)...))))))))((((.....))))........)).)..))))).)..--- ( -24.90, z-score = -1.71, R) >droPer1.super_8 3500188 105 - 3966273 CAUUGCGGCUUGCAAACGACCAAGUUAUGAUAAUGUUUGGCCAACUAAACUCGUUUA-GCCUGCCAAAUAGUAGUACGACUGCUCCCCCAUUCCUCCCUUGGCCAU--- ..((((.....))))..(.(((((..(.((...((((((((...(((((....))))-)...))))))))((((.....))))........)).)..))))).)..--- ( -24.90, z-score = -1.71, R) >droWil1.scaffold_180772 6780336 90 + 8906247 CAUUGCGGCUUGCAAACGACCAAGUUAUGAUAAUGUUUGGCCAACUAAACUGGUUUAGGCUUGCCAAAUAUUGCCACCACCUAAAACGAA------------------- ......(((.....(((......))).....((((((((((...(((((....)))))....)))))))))))))...............------------------- ( -20.60, z-score = -0.88, R) >droMoj3.scaffold_6500 8672422 97 + 32352404 CAUUGCGGCUUUGAAACGACCAAGUUAUGAUAAUGUUUGGCCAACUAAACUCGUUUA-GCUUGCCAAAUACGAAGCCCACAAAAAGAACUGGCUCGGU----------- ...((.(((((((.(((......))).......((((((((...(((((....))))-)...)))))))))))))))))...................----------- ( -24.00, z-score = -1.44, R) >droGri2.scaffold_15252 7377026 106 - 17193109 CAUUGCGGCUUGCAAACGACCAAGUUAUGAUAAUGUUUGGCCAACUAAACUCGUUUA-GCUUGCCAAAUACGAAGCCCACAAAAACUGCACACCCAAAUCGAGUAUU-- ..(((.((..(((((((......))).......((((((((...(((((....))))-)...))))))))................))))..)))))..........-- ( -21.00, z-score = -0.88, R) >droVir3.scaffold_12963 19527690 81 - 20206255 CAUUGCGGCUUGCAAACGACCAAGUUAUGAUAAUGUUUGGCCAACUAAACUCGUUUA-CCCAGCCAAAUACGAAGCCCACAG--------------------------- ...((.(((((.(.(((......))).......((((((((....((((....))))-....)))))))).))))))))...--------------------------- ( -17.20, z-score = -1.54, R) >consensus CAUUGCGGCUUGCAAACGACCAAGUUAUGAUAAUGUUUGGCCAACUAAACUCGUUUA_GCUUGCCAAAUACGACUGCCAC________AACCCCAGAAACUCCC_CU__ .((((..(((((........)))))..))))..((((((((....((((....)))).....))))))))....................................... (-14.65 = -14.75 + 0.09)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:07:24 2011