
| Sequence ID | dm3.chrX |
|---|---|
| Location | 9,847,739 – 9,847,802 |
| Length | 63 |
| Max. P | 0.999175 |

| Location | 9,847,739 – 9,847,802 |
|---|---|
| Length | 63 |
| Sequences | 5 |
| Columns | 72 |
| Reading direction | forward |
| Mean pairwise identity | 87.08 |
| Shannon entropy | 0.22859 |
| G+C content | 0.39216 |
| Mean single sequence MFE | -19.02 |
| Consensus MFE | -13.14 |
| Energy contribution | -13.94 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.776571 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
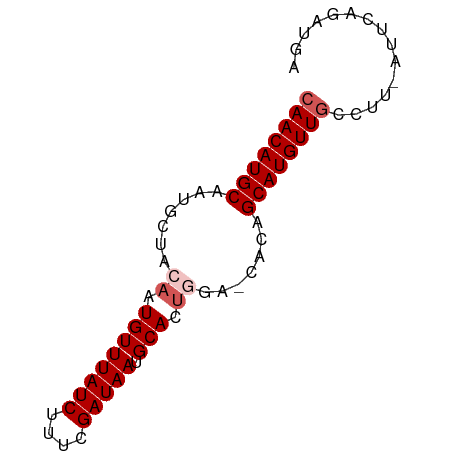
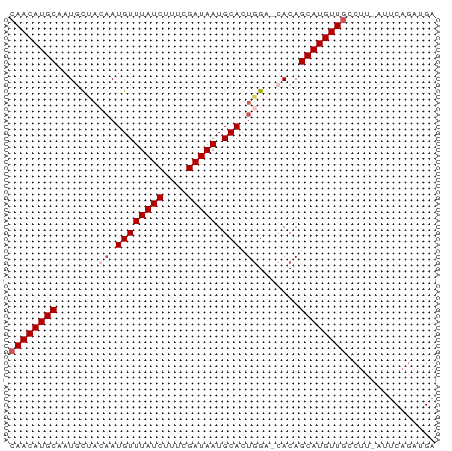
>dm3.chrX 9847739 63 + 22422827 CAACAUGCAAUGCUACAAUGUUUAUCUUUCGAUAAUGCACUGGAACACAGCAUGUU---------CAGAUGA .(((((((..((...((.((((((((....))))).))).))...))..)))))))---------....... ( -14.40, z-score = -1.35, R) >droSim1.chrX 7846392 71 + 17042790 CAACAUGCAAUGCUACAAUGUUUAUCUUUCGAUAAUGCACUGGAGCACAGCAUGUUGCCUU-AUUCAGAUGA ((((((((..((((.((.((((((((....))))).))).)).))))..))))))))....-.......... ( -22.30, z-score = -3.04, R) >droSec1.super_15 612401 71 + 1954846 CAACAUGCAAUGCUACAAUGUUUAUCUUUCGAUAAUGCACUGGAGCACAGCAUGUUGCCUU-AUUCAGAUGA ((((((((..((((.((.((((((((....))))).))).)).))))..))))))))....-.......... ( -22.30, z-score = -3.04, R) >droYak2.chrX 18441031 70 + 21770863 CAACAUGCCAUCUAUGCAUGUUUAUCUUUCGAUAAUGCACUGG--CACAGCAUGUUGCCUUCAUUCAGAUGA ((((((((...((((((((...((((....))))))))).)))--....))))))))...((((....)))) ( -17.30, z-score = -1.18, R) >droEre2.scaffold_4690 13485763 69 - 18748788 CAACAUGCAAUGCUAGAAUGUUUAUCGUUCGAUAAUGCACUAG--AACAGCAUGUUGCCUU-AUUCAGAUGA ((((((((....((((..((((((((....))))).)))))))--....))))))))....-.......... ( -18.80, z-score = -2.32, R) >consensus CAACAUGCAAUGCUACAAUGUUUAUCUUUCGAUAAUGCACUGGA_CACAGCAUGUUGCCUU_AUUCAGAUGA ((((((((.......((.((((((((....))))).))).)).......))))))))............... (-13.14 = -13.94 + 0.80)

| Location | 9,847,739 – 9,847,802 |
|---|---|
| Length | 63 |
| Sequences | 5 |
| Columns | 72 |
| Reading direction | reverse |
| Mean pairwise identity | 87.08 |
| Shannon entropy | 0.22859 |
| G+C content | 0.39216 |
| Mean single sequence MFE | -25.24 |
| Consensus MFE | -21.66 |
| Energy contribution | -23.58 |
| Covariance contribution | 1.92 |
| Combinations/Pair | 1.04 |
| Mean z-score | -4.27 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.69 |
| SVM RNA-class probability | 0.999175 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
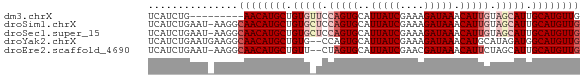
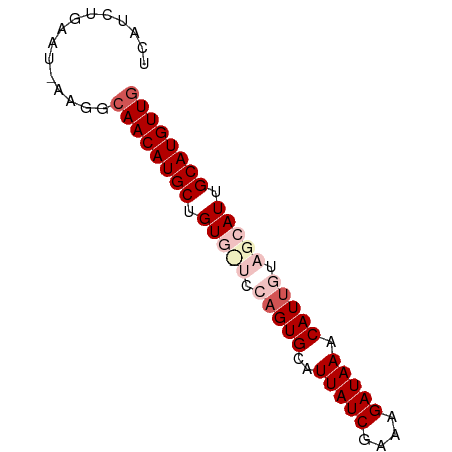
>dm3.chrX 9847739 63 - 22422827 UCAUCUG---------AACAUGCUGUGUUCCAGUGCAUUAUCGAAAGAUAAACAUUGUAGCAUUGCAUGUUG .......---------(((((((.(((((.(((((..(((((....))))).))))).))))).))))))). ( -25.40, z-score = -5.19, R) >droSim1.chrX 7846392 71 - 17042790 UCAUCUGAAU-AAGGCAACAUGCUGUGCUCCAGUGCAUUAUCGAAAGAUAAACAUUGUAGCAUUGCAUGUUG ..........-....((((((((.(((((.(((((..(((((....))))).))))).))))).)))))))) ( -29.50, z-score = -5.47, R) >droSec1.super_15 612401 71 - 1954846 UCAUCUGAAU-AAGGCAACAUGCUGUGCUCCAGUGCAUUAUCGAAAGAUAAACAUUGUAGCAUUGCAUGUUG ..........-....((((((((.(((((.(((((..(((((....))))).))))).))))).)))))))) ( -29.50, z-score = -5.47, R) >droYak2.chrX 18441031 70 - 21770863 UCAUCUGAAUGAAGGCAACAUGCUGUG--CCAGUGCAUUAUCGAAAGAUAAACAUGCAUAGAUGGCAUGUUG ((((....))))...(((((((((((.--(..((((((((((....))))...)))))).)))))))))))) ( -23.80, z-score = -3.06, R) >droEre2.scaffold_4690 13485763 69 + 18748788 UCAUCUGAAU-AAGGCAACAUGCUGUU--CUAGUGCAUUAUCGAACGAUAAACAUUCUAGCAUUGCAUGUUG ..........-....((((((((.((.--((((....(((((....))))).....)))).)).)))))))) ( -18.00, z-score = -2.19, R) >consensus UCAUCUGAAU_AAGGCAACAUGCUGUG_UCCAGUGCAUUAUCGAAAGAUAAACAUUGUAGCAUUGCAUGUUG ...............((((((((.(((((.(((((..(((((....))))).))))).))))).)))))))) (-21.66 = -23.58 + 1.92)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:29:54 2011