| Sequence ID | dm3.chrX |
|---|---|
| Location | 9,844,618 – 9,844,717 |
| Length | 99 |
| Max. P | 0.658484 |

| Location | 9,844,618 – 9,844,717 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 58.87 |
| Shannon entropy | 0.79777 |
| G+C content | 0.49905 |
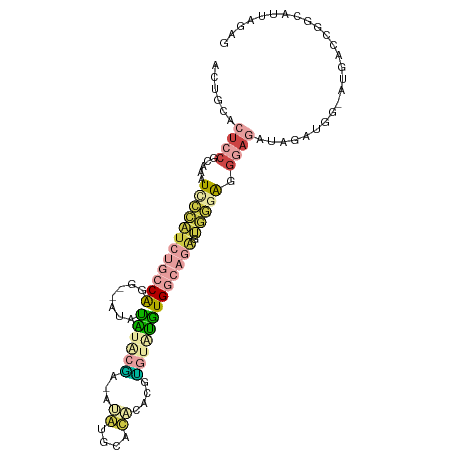
| Mean single sequence MFE | -30.90 |
| Consensus MFE | -9.58 |
| Energy contribution | -11.13 |
| Covariance contribution | 1.55 |
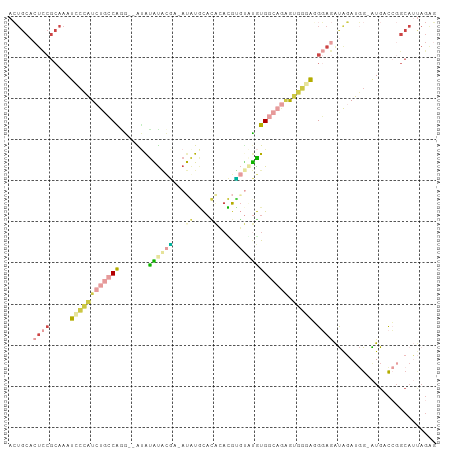
| Combinations/Pair | 1.75 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.31 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.658484 |
| Prediction | RNA |
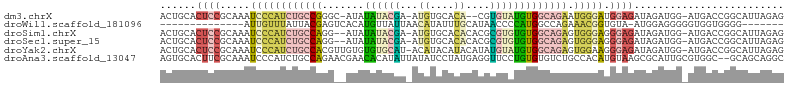
Download alignment: ClustalW | MAF
>dm3.chrX 9844618 99 + 22422827 ACUGCACUCCGCAAAUCCCAUCUGCCGGGC-AUAUAUACGA-AUGUGCACA--CGUGUAUGUGGCAGAAUGGGAUGGAGAUAGAUGG-AUGACCGGCAUUAGAG ..(((.((((....((((((((((((....-(((((((((.-.((....))--)))))))))))))).))))))))))).....(((-....))))))...... ( -34.50, z-score = -2.17, R) >droWil1.scaffold_181096 9360474 82 - 12416693 --------------AUUGUUUAUUACGAGUCACAUGUUAUUAACAUAUUUGCAUAACCCCAUGGCCAGAAACGGUGUA-AUGGAGGGGGUGGUGGGG------- --------------.((((.....)))).((((((((.....)))).....(((..((((((.(((......)))...-)))).))..)))))))..------- ( -18.80, z-score = 0.44, R) >droSim1.chrX 7843292 100 + 17042790 ACUGCACUCCGCAAAUCCCAUCUGCCAGG--AUAUAUACGA-AUGUGCACACACGCGUGUGUGGCAGAGUGGGAGGGAGAUAGAUGG-AUGACCGGCAUUAGAG ..(((.((((.....((((((((((((..--...((((((.-.(((....)))..))))))))))))).))))).)))).....(((-....))))))...... ( -35.60, z-score = -2.25, R) >droSec1.super_15 609294 100 + 1954846 ACUGCACUCCGCAAAUCCCAUCUGCCAGG--AUAUAUACGA-AUGUGCACACACGCGUGUGUGGCAGAGUGGGAGGGAGAUAGAUGG-AUGACCGGCAUUAGAG ..(((.((((.....((((((((((((..--...((((((.-.(((....)))..))))))))))))).))))).)))).....(((-....))))))...... ( -35.60, z-score = -2.25, R) >droYak2.chrX 18437778 102 + 21770863 ACUGCACUCCGCAAAUCCCAUCUGCCACGUUGUGUGUGCAU-ACAUACAUACAUAUGUAUGUGGCAGAGUGGAAGGGAGAUAGAUGG-AUGACCGGCAUUAGAG ..(((.((((.(...(((..((((((((((((((((((...-.....))))))))...))))))))))..))).))))).....(((-....))))))...... ( -35.90, z-score = -2.41, R) >droAna3.scaffold_13047 221900 102 + 1816235 AGUGCACUUCGCAAAUCCCAUCUGCCAGAACGAACACAUAUUAUAUCCUAUGAGGUUCCUGUGUGUCUGCCACAUGUAAGCGCAUUGCGUGGC--GCAGCAGGC ..(((.....)))....((..((((......((.((((((......((.....))....)))))))).(((((..((((.....)))))))))--))))..)). ( -25.00, z-score = 1.05, R) >consensus ACUGCACUCCGCAAAUCCCAUCUGCCAGG__AUAUAUACGA_AUAUGCACACACGUGUAUGUGGCAGAGUGGGAGGGAGAUAGAUGG_AUGACCGGCAUUAGAG ......((((.....((((((((((((.......((((((...((....))....))))))))))))).))))).))))......................... ( -9.58 = -11.13 + 1.55)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:29:52 2011