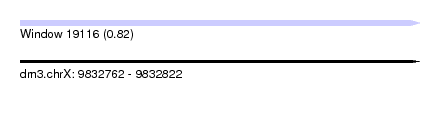
| Sequence ID | dm3.chrX |
|---|---|
| Location | 9,832,762 – 9,832,822 |
| Length | 60 |
| Max. P | 0.817701 |

| Location | 9,832,762 – 9,832,822 |
|---|---|
| Length | 60 |
| Sequences | 12 |
| Columns | 68 |
| Reading direction | forward |
| Mean pairwise identity | 78.55 |
| Shannon entropy | 0.43974 |
| G+C content | 0.50548 |
| Mean single sequence MFE | -17.62 |
| Consensus MFE | -8.94 |
| Energy contribution | -9.11 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.817701 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 9832762 60 + 22422827 --GACUUUGGGGUAUGACUUAUUCAUGUCAACCGCUGACAAACGGCGUCGAGAGCUGCAGCU------ --(((.....(((.((((........)))))))((((.....)))))))...(((....)))------ ( -16.50, z-score = -0.92, R) >droSim1.chrX 7831490 60 + 17042790 --GACUUUGGGGUAUGACUUAUUCAUGUCAACCGCUGACAAACGGCGUCGAGAGCUGCAGAU------ --...((((.(((.((((........))))..(((((.....)))))......))).)))).------ ( -15.10, z-score = -0.72, R) >droSec1.super_15 597209 60 + 1954846 --GACUUUGGGGUAUGACUUAUUCAUGUCAACCGCUGACAAACGGCGUCGAGAGCUGCAGAU------ --...((((.(((.((((........))))..(((((.....)))))......))).)))).------ ( -15.10, z-score = -0.72, R) >droYak2.chrX 18425812 60 + 21770863 --GACUUUGGGGUAUGACUUAUUCAUGUCAACCGCUGACAAACGGCGUCGAGAGCUGCCGAU------ --....(..((((.((((........))))))).)..)....(((((.(....).)))))..------ ( -18.50, z-score = -1.83, R) >droEre2.scaffold_4690 13469702 60 - 18748788 --GACUUUGGGGUAUGACUUAUUCAUGUCAACCGCUGACAAACGGCGUCGAGAGCUGCAGAU------ --...((((.(((.((((........))))..(((((.....)))))......))).)))).------ ( -15.10, z-score = -0.72, R) >droAna3.scaffold_13047 1547714 57 - 1816235 ---GGCUCGGGCUAUGACUUAUUCAUGUCAACCGCUGACAAACGGCGU--GGGCCUGCAGAU------ ---..((((((((.((((........)))).((((((.....))))).--))))))).))..------ ( -20.10, z-score = -2.03, R) >dp4.chrXL_group3a 1612476 57 - 2690836 ---ACUUUUGGCUAUGACUUAUUCAUGUCAACCGCUGACAAACGGCGGC--CAACUGCAGAU------ ---....(((((..((((........))))..(((((.....)))))))--)))........------ ( -16.60, z-score = -2.89, R) >droPer1.super_15 1814099 57 + 2181545 ---ACUUUUGGCUAUGACUUAUUCAUGUCAACCGCUGACAAACGGCGGC--CAACUGCAGAU------ ---....(((((..((((........))))..(((((.....)))))))--)))........------ ( -16.60, z-score = -2.89, R) >droWil1.scaffold_181096 9347550 62 - 12416693 AACUUUCCUGGCCAUGACUUAUUCAUGUCAACCGCUGACAAACGGCUUGUCGAGCUGGAAAU------ ...(((((((((.((((.....))))))))...((((((((.....))))).))).))))).------ ( -17.10, z-score = -2.45, R) >droVir3.scaffold_12970 2955747 50 - 11907090 --GCCUGUUGGCUAUGACUUAUUCAUGUCAGCCGCUAACA--CGGCUC---GAGUUG----------- --((((((((((..((((........))))...)))))))--.)))..---......----------- ( -19.10, z-score = -3.60, R) >droMoj3.scaffold_6473 16531526 57 + 16943266 --GCCUGUUGGCUAUGACUUAUUCAUGUCAGCCGCUGACAAACGGCUC---GAGUCGGAGCU------ --...(((..((..((((........))))...))..)))...(((((---......)))))------ ( -18.80, z-score = -1.51, R) >droGri2.scaffold_15203 352279 63 - 11997470 --UGCUGCUGUCUAUGACUUAUUCAUGUCAGCCGCUGACAAACGGCUC---GAGUUGAAUCAGCAGCU --.(((((((..........(((((..(((((((........))))).---))..)))))))))))). ( -22.90, z-score = -2.61, R) >consensus __GACUUUGGGCUAUGACUUAUUCAUGUCAACCGCUGACAAACGGCGUC__GAGCUGCAGAU______ .........((...((((........)))).))((((.....))))...................... ( -8.94 = -9.11 + 0.17)

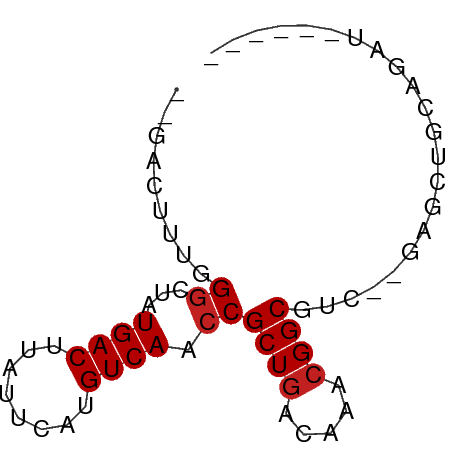
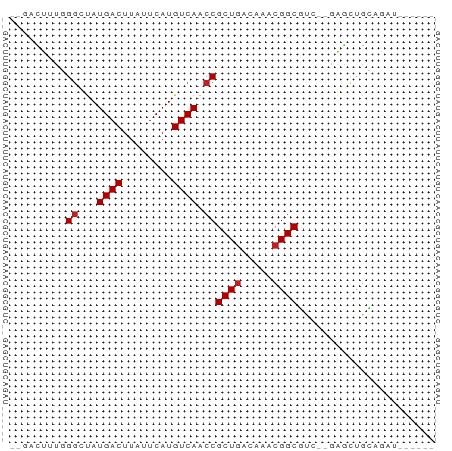
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:29:49 2011