| Sequence ID | dm3.chrX |
|---|---|
| Location | 9,829,353 – 9,829,443 |
| Length | 90 |
| Max. P | 0.825242 |

| Location | 9,829,353 – 9,829,443 |
|---|---|
| Length | 90 |
| Sequences | 9 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 66.88 |
| Shannon entropy | 0.66736 |
| G+C content | 0.58798 |
| Mean single sequence MFE | -27.26 |
| Consensus MFE | -13.83 |
| Energy contribution | -13.51 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.38 |
| Mean z-score | -0.83 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.825242 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
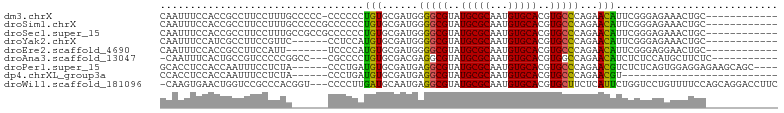
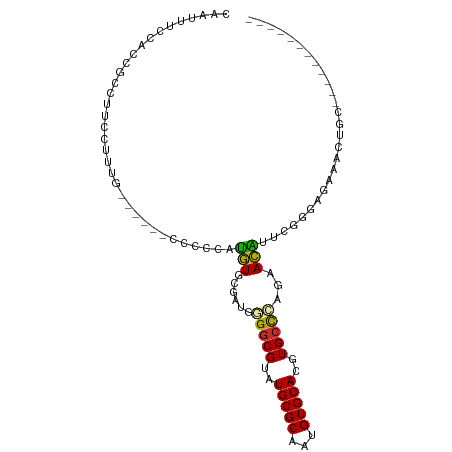
>dm3.chrX 9829353 90 - 22422827 CAAUUUCCACCGCCUUCCUUUGCCCCC-CCCCCCUGUGCGAUGGGGCGUAUGCGCAAUGUGCACGUGCCCAGAACAUUCGGGAGAAACUGC------------ ((.((((..(((..((((.((((....-.........)))).)(((((..(((((...)))))..))))).)))....)))..)))).)).------------ ( -24.72, z-score = 0.03, R) >droSim1.chrX 7828177 91 - 17042790 CAAUUUCCACCGCCUUCCUUUGCCCCCGCCCCCCUGUGCGAUGGGGCGUAUGCGCAAUGUGCACGUGCCCAGAACAUUCGGGAGAAACUGC------------ ((.((((..(((..(((....((((((((........)))..)))))(((((.((.....)).)))))...)))....)))..)))).)).------------ ( -28.00, z-score = -0.58, R) >droSec1.super_15 593910 91 - 1954846 CAAUUUCCACCGCCUUCCUUUGCCGCCGCCCCCCUGUGCGAUGGGGCGUAUGCGCAAUGUGCACGUGCCCAGAACAUUCGGGAGAAACUGC------------ ((.((((..(((..(((......(((((......)).)))...(((((..(((((...)))))..))))).)))....)))..)))).)).------------ ( -27.00, z-score = 0.26, R) >droYak2.chrX 18422213 85 - 21770863 CAAUUUCCAUCGCCUUCCGUUC------CCUCCAUGUGCGAUGGGGCGUAUGCGCAAUGUGCACGUGCCCAGAACAUUCGGGAGAAACUGC------------ ...(((((((((((........------.......).))))).(((((..(((((...)))))..)))))..........)))))......------------ ( -26.36, z-score = -0.55, R) >droEre2.scaffold_4690 13466429 84 + 18748788 CAAUUUCCACCGCCUUCCAUU-------UCCCCAUGUGCGAUGGGGCGUAUGCGCAAUGUGCACGUGCCCAGAACAUUCGGGAGGAACUGC------------ ...........((.((((...-------((((.((((.(....(((((..(((((...)))))..))))).).))))..))))))))..))------------ ( -28.10, z-score = -0.87, R) >droAna3.scaffold_13047 1545113 88 + 1816235 -CAAUUUCACUGCCGUCCCCCGGCC---CGCCCCUGUGCGACGAGGCGUAUGCGCAAUGUGCACGUGGCCAGAACAUCUCUCCAUGCUUCUC----------- -..........((((.....)))).---.(((..(((((.(((..(((....)))..)))))))).))).(((((((......))).)))).----------- ( -24.10, z-score = -0.16, R) >droPer1.super_15 349697 93 + 2181545 GCACCUCCACCAAUUUCCUCUA------CCCUGAUGUGCGAUGAGGCGUAUGCGCAAUGUGCACGUGCCCAGAACGUCUCUCAGUGGAGGAGAAGCAGC---- .............(((((((((------(...(((((....((.((((..(((((...)))))..))))))..))))).....))))))))))......---- ( -33.90, z-score = -2.25, R) >dp4.chrXL_group3a 1060780 71 - 2690836 CCACCUCCACCAAUUUCCUCUA------CCCUGAUGUGCGAUGAGGCGUAUGCGCAAUGUGCACGUGCCCAGAACGU-------------------------- ......................------.....((((....((.((((..(((((...)))))..))))))..))))-------------------------- ( -16.00, z-score = -0.44, R) >droWil1.scaffold_181096 9338202 99 + 12416693 -CAAGUGAACUGGUCCGCCCACGGU---CCCCUUGAUGCAAUGAGGCGUAUGCGCAAUGUGCACGUGCUUCUCAUUCUGGUCCUGUUUUCCAGCAGGACCUUC -..(((((...((.(((....))).---))............((((((..(((((...)))))..)))))))))))..((((((((......))))))))... ( -37.20, z-score = -2.86, R) >consensus CAAUUUCCACCGCCUUCCUUUG______CCCCCAUGUGCGAUGGGGCGUAUGCGCAAUGUGCACGUGCCCAGAACAUUCGGGAGAAACUGC____________ ..................................(((......(((((..(((((...)))))..)))))...)))........................... (-13.83 = -13.51 + -0.32)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:29:48 2011