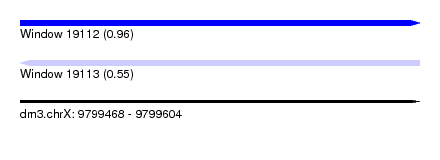
| Sequence ID | dm3.chrX |
|---|---|
| Location | 9,799,468 – 9,799,604 |
| Length | 136 |
| Max. P | 0.962170 |

| Location | 9,799,468 – 9,799,604 |
|---|---|
| Length | 136 |
| Sequences | 5 |
| Columns | 139 |
| Reading direction | forward |
| Mean pairwise identity | 47.67 |
| Shannon entropy | 0.94115 |
| G+C content | 0.37745 |
| Mean single sequence MFE | -37.06 |
| Consensus MFE | -6.12 |
| Energy contribution | -7.12 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.96 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.17 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.962170 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
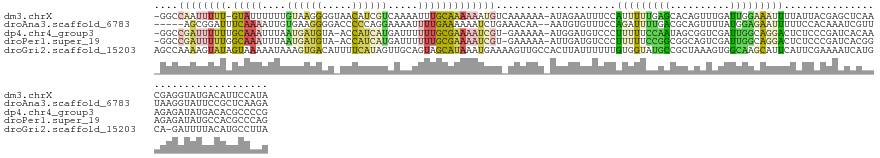
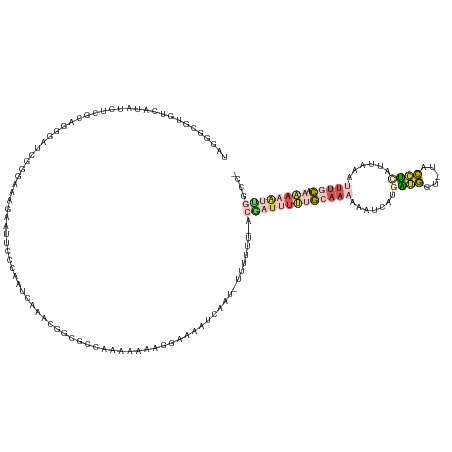
>dm3.chrX 9799468 136 + 22422827 -GGCCAAUUUUU-GUAUUUUUUGUAAGGGGUAACAUCGUCAAAAUUUGCAAAAAAUGUCAAAAAA-AUAGAAUUUCCAUUUUUGAGCACAGUUUGAUUGGAAAUUUUAUUACGAGCUCAACGAGGUAUGACAUUCCAUA -((....(((((-(((..(((((...(((....).))..)))))..))))))))((((((....(-((((((((((((...(..(((...)))..).)))))))))))))...(.(((...))).).)))))).))... ( -32.10, z-score = -2.45, R) >droAna3.scaffold_6783 2871 132 - 5528 -----AGCGGAUUUCAAAAUUGUGAAGGGGACCCCCAGGAAAAUUUUGAAAAAAUCUGAAACAA--AAUGUGUUUCCAGAUUUUGACGCAGUUUUAUGGAGAAUUUUUCCACAAAUCGUUUAAGGUAUUCCGCUCAAGA -----((((((..(((((((((((..(((....)))..............(((((((((((((.--....))))).))))))))..))))))))).((((((...))))))............))...))))))..... ( -37.80, z-score = -2.68, R) >dp4.chr4_group3 844715 135 + 11692001 -GGCCGAUUUUUUGCAAAUUUAAUGAUGUA-ACCAUCAUGAUUUUUUGCGAAAAUCGU-GAAAAA-AUGGAUGUCCCUUUUUCCAAUAGCGGUCGAUUGGCAGGACUCUCCCGAUCACAAAGAGAUAUGACACGCCCCG -((.((((((((.(((((....((((((..-..)))))).....)))))))))))))(-(.((((-(.((....)).))))).))...((((((((((((.((....)).)))))).(.....)....))).))).)). ( -35.50, z-score = -1.73, R) >droPer1.super_19 1829929 135 + 1869541 -GGCCGAUUUUUGGCAAAUUUAAUGAUGUA-ACCAUCAUGAUUUUUUGCGAAAAUCGU-GAAAAA-AUUGAUGUCCCUUUUUCCGGCGGCAGUCGAUUGGCAGGACUCUCCCGAUCACGGAGAGAUAUGCCACGCCCAG -((((((((((((.((((....((((((..-..)))))).....))))))))))))).-.....(-(((((((((((.......)).)))).))))))((((...((((((.......))))))...))))..)))... ( -46.50, z-score = -3.34, R) >droGri2.scaffold_15203 315003 138 - 11997470 AGCCAAAAGUAUAGUAAAAAUAAAGUGACAUUUUCAUAGUUGCAGUAGCAUAAAUGAAAAGUUGCCACUUAUUUUUUGUGGUAUGCCGCUAAAGUGGCAAGCAUUCAUUCGAAAAUCAUGCA-GAUUUUACAUGCCUUA ........((((.((((((.....((((..(((((.....(((....)))..((((((...(((((((((.......((((....))))..)))))))))...)))))).)))))))))...-..)))))))))).... ( -33.40, z-score = -1.91, R) >consensus _GGCCAAUUUUUUGCAAAUUUAAUGAGGGA_ACCAUCAUGAAAUUUUGCAAAAAUCGU_AAAAAA_AUUGAUUUUCCAUUUUCCGACGCAGGUCGAUUGGCAAUUCUCUCACGAACACAAAGAGAUAUGACACGCCAUA ....((((((((.(((((....((((((.....)))))).....)))))))))))))....................(((((((((..........))))))))).................................. ( -6.12 = -7.12 + 1.00)

| Location | 9,799,468 – 9,799,604 |
|---|---|
| Length | 136 |
| Sequences | 5 |
| Columns | 139 |
| Reading direction | reverse |
| Mean pairwise identity | 47.67 |
| Shannon entropy | 0.94115 |
| G+C content | 0.37745 |
| Mean single sequence MFE | -35.20 |
| Consensus MFE | -1.82 |
| Energy contribution | -1.26 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 2.25 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.05 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.546537 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
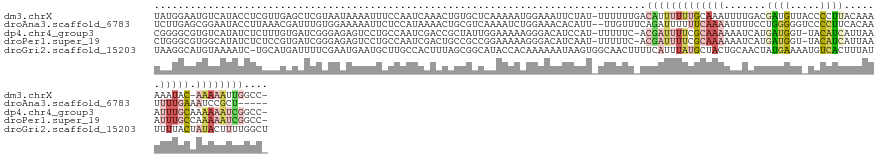
>dm3.chrX 9799468 136 - 22422827 UAUGGAAUGUCAUACCUCGUUGAGCUCGUAAUAAAAUUUCCAAUCAAACUGUGCUCAAAAAUGGAAAUUCUAU-UUUUUUGACAUUUUUUGCAAAUUUUGACGAUGUUACCCCUUACAAAAAAUAC-AAAAAUUGGCC- ...(((((((((.........((((.((.....................)).))))((((((((.....))))-)))).)))))))))...(((.(((((....(((........))).......)-)))).)))...- ( -18.90, z-score = 0.33, R) >droAna3.scaffold_6783 2871 132 + 5528 UCUUGAGCGGAAUACCUUAAACGAUUUGUGGAAAAAUUCUCCAUAAAACUGCGUCAAAAUCUGGAAACACAUU--UUGUUUCAGAUUUUUUCAAAAUUUUCCUGGGGGUCCCCUUCACAAUUUUGAAAUCCGCU----- .....((((((.............((((((((.......)))))))).(((...((((((.((....)).)))--)))...)))....((((((((((.....(((....))).....))))))))))))))))----- ( -39.60, z-score = -4.32, R) >dp4.chr4_group3 844715 135 - 11692001 CGGGGCGUGUCAUAUCUCUUUGUGAUCGGGAGAGUCCUGCCAAUCGACCGCUAUUGGAAAAAGGGACAUCCAU-UUUUUC-ACGAUUUUCGCAAAAAAUCAUGAUGGU-UACAUCAUUAAAUUUGCAAAAAAUCGGCC- .((((((.((((((......)))(((.((.((....)).)).))))))))))..((((((((.((....)).)-))))))-)(((((((.(((((.....((((((..-..))))))....)))))..))))))).))- ( -38.40, z-score = -1.85, R) >droPer1.super_19 1829929 135 - 1869541 CUGGGCGUGGCAUAUCUCUCCGUGAUCGGGAGAGUCCUGCCAAUCGACUGCCGCCGGAAAAAGGGACAUCAAU-UUUUUC-ACGAUUUUCGCAAAAAAUCAUGAUGGU-UACAUCAUUAAAUUUGCCAAAAAUCGGCC- ...(((((((((...((((((.......))))))...)))))......))))((((((((((..(....)..)-))))))-..((((((.(((((.....((((((..-..))))))....)))))..))))))))).- ( -45.40, z-score = -3.03, R) >droGri2.scaffold_15203 315003 138 + 11997470 UAAGGCAUGUAAAAUC-UGCAUGAUUUUCGAAUGAAUGCUUGCCACUUUAGCGGCAUACCACAAAAAAUAAGUGGCAACUUUUCAUUUAUGCUACUGCAACUAUGAAAAUGUCACUUUAUUUUUACUAUACUUUUGGCU ..((((((..((((((-.....)))))).......))))))((((.....(.((....)).).((((((((((((((..(((((((...(((....)))...))))))))))))).))))))))..........)))). ( -33.70, z-score = -3.05, R) >consensus UAGGGCGUGUCAUAUCUCGCAGGGAUCGGGAAAGAAUUCCCAAUCAAACGGCGCCAAAAAAAGGAAAAUCAAU_UUUUUU_ACGAUUUUUGCAAAAAAUCAUGAUGGU_UACCUCAUUAAAUUUGCAAAAAAUUGGCC_ ......................................................................................................((((.....))))........................ ( -1.82 = -1.26 + -0.56)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:29:46 2011