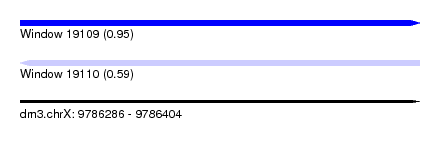
| Sequence ID | dm3.chrX |
|---|---|
| Location | 9,786,286 – 9,786,404 |
| Length | 118 |
| Max. P | 0.951678 |

| Location | 9,786,286 – 9,786,404 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.42 |
| Shannon entropy | 0.48144 |
| G+C content | 0.38156 |
| Mean single sequence MFE | -27.29 |
| Consensus MFE | -12.96 |
| Energy contribution | -13.04 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.951678 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
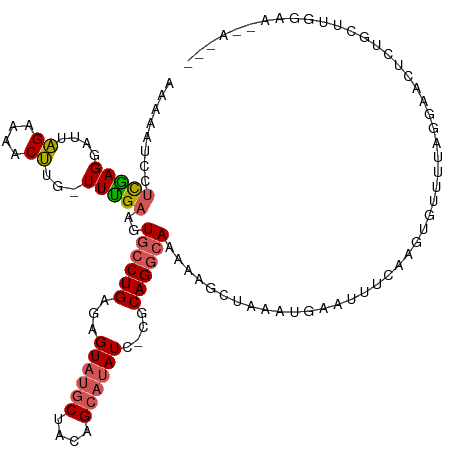
>dm3.chrX 9786286 118 + 22422827 UAGUUCCCAAGAGCAGAAUUCCUAAAACACUUGAAAUUCAUUUAGCUUUUUGCCUGCA-GAUAUGCUGUAGCAUACUCACAGGCACUUCAAA-AAAGUUUUCUAAUCCUUGAGGAUUUUU ....(((((((....(((((.(..........).)))))....(((((((((((((..-(((((((....))))).)).))))))......)-))))))........)))).)))..... ( -25.90, z-score = -1.48, R) >droEre2.scaffold_4690 13417469 113 - 18748788 ------UGCCAAGCUGAGUUUUUAGAGCACUUGAAAUUCAUUUAGCUUUCUGCCUGCGCAAUAUUCUGUUGCAUACUCUCAGGCAGUUCGAA-AAAGUUUUCUAAUCCUCAGACAUUUCC ------......((((((...((((((.(((((.....)..........(((((((.(((((.....))))).......)))))))......-.)))).))))))..))))).)...... ( -26.00, z-score = -1.20, R) >droYak2.chrX 18378386 99 + 21770863 -------------------UUCCAAAUCACUUGAAAUUCAUAUAGCUUUCUGCCUGCG-GAUAAGCUGUAGCAAACUUUCAGUUACUUCAAA-CUGGUUUUCCAAUCCUCGAGGAUCAUU -------------------......(((.(((((......((((((((((((....))-)).))))))))........((((((......))-))))...........)))))))).... ( -22.70, z-score = -2.42, R) >droSec1.super_15 546786 116 + 1954846 ---UAGUUCCAUGCAGAGUUCGUAAAACGCUUGAAAUUCAUUUAGCUUUUUGCCUGCG-GAUAUGCUGUAGCAUACUCUCAGGCACUUCAAAACAAGUUUUCUAAUUCUCGAGGAUUUUU ---....(((.((.((((((.(.((((((((.((......)).)))....((((((.(-..(((((....)))))..).))))))...........)))))).)))))))).)))..... ( -31.60, z-score = -2.80, R) >droSim1.chrX 7789053 105 + 17042790 ---UAGUUCCACGCAGAGUUC-----------GAAAUUCAUUUAGCUUUUUGCCUGCG-GAUAUGCUGUAGCAUACUCUCAGGCACUUCAAAACAAGUUUUCUAAUUCUCGAGGAUUUUU ---....(((.((.((((((.-----------((((..............((((((.(-..(((((....)))))..).)))))).............)))).)))))))).)))..... ( -30.23, z-score = -3.52, R) >consensus ___U__UUCCAAGCAGAGUUCCUAAAACACUUGAAAUUCAUUUAGCUUUUUGCCUGCG_GAUAUGCUGUAGCAUACUCUCAGGCACUUCAAA_CAAGUUUUCUAAUCCUCGAGGAUUUUU ..................................................((((((.....(((((....)))))....)))))).........((((((((........)))))))).. (-12.96 = -13.04 + 0.08)


| Location | 9,786,286 – 9,786,404 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.42 |
| Shannon entropy | 0.48144 |
| G+C content | 0.38156 |
| Mean single sequence MFE | -27.88 |
| Consensus MFE | -12.74 |
| Energy contribution | -13.14 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.589390 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 9786286 118 - 22422827 AAAAAUCCUCAAGGAUUAGAAAACUUU-UUUGAAGUGCCUGUGAGUAUGCUACAGCAUAUC-UGCAGGCAAAAAGCUAAAUGAAUUUCAAGUGUUUUAGGAAUUCUGCUCUUGGGAACUA .....(((.(((((.((((...(((((-...)))))((((((..((((((....)))))).-.))))))......))))..(((((((..........)))))))...)))))))).... ( -34.60, z-score = -2.81, R) >droEre2.scaffold_4690 13417469 113 + 18748788 GGAAAUGUCUGAGGAUUAGAAAACUUU-UUCGAACUGCCUGAGAGUAUGCAACAGAAUAUUGCGCAGGCAGAAAGCUAAAUGAAUUUCAAGUGCUCUAAAAACUCAGCUUGGCA------ ......(.(((((..(((((..(((..-......(((((((.((((((........))))).).))))))).........((.....)))))..)))))...))))))......------ ( -27.10, z-score = -0.46, R) >droYak2.chrX 18378386 99 - 21770863 AAUGAUCCUCGAGGAUUGGAAAACCAG-UUUGAAGUAACUGAAAGUUUGCUACAGCUUAUC-CGCAGGCAGAAAGCUAUAUGAAUUUCAAGUGAUUUGGAA------------------- .....(((....(((((((....))))-))).((((.(((..(((((((.((.(((((.((-.(....).))))))).)))))))))..))).))))))).------------------- ( -19.30, z-score = 0.22, R) >droSec1.super_15 546786 116 - 1954846 AAAAAUCCUCGAGAAUUAGAAAACUUGUUUUGAAGUGCCUGAGAGUAUGCUACAGCAUAUC-CGCAGGCAAAAAGCUAAAUGAAUUUCAAGCGUUUUACGAACUCUGCAUGGAACUA--- .....(((..((((.((..(((((..(((((....((((((.(..(((((....)))))..-).)))))).)))))....((.....))...)))))...)).))).)..)))....--- ( -28.60, z-score = -1.85, R) >droSim1.chrX 7789053 105 - 17042790 AAAAAUCCUCGAGAAUUAGAAAACUUGUUUUGAAGUGCCUGAGAGUAUGCUACAGCAUAUC-CGCAGGCAAAAAGCUAAAUGAAUUUC-----------GAACUCUGCGUGGAACUA--- .....(((.(((((.((.((((..(..(((.(...((((((.(..(((((....)))))..-).)))))).....).)))..).))))-----------.)).))).)).)))....--- ( -29.80, z-score = -3.26, R) >consensus AAAAAUCCUCGAGGAUUAGAAAACUUG_UUUGAAGUGCCUGAGAGUAUGCUACAGCAUAUC_CGCAGGCAAAAAGCUAAAUGAAUUUCAAGUGUUUUAGGAACUCUGCUUGGAA__A___ ........(((((....((....))...)))))..((((((...((((((....))))))....)))))).................................................. (-12.74 = -13.14 + 0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:29:43 2011