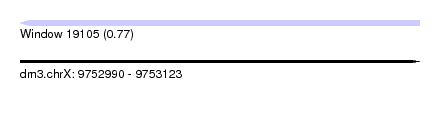
| Sequence ID | dm3.chrX |
|---|---|
| Location | 9,752,990 – 9,753,123 |
| Length | 133 |
| Max. P | 0.766863 |

| Location | 9,752,990 – 9,753,123 |
|---|---|
| Length | 133 |
| Sequences | 5 |
| Columns | 156 |
| Reading direction | reverse |
| Mean pairwise identity | 65.58 |
| Shannon entropy | 0.60188 |
| G+C content | 0.52604 |
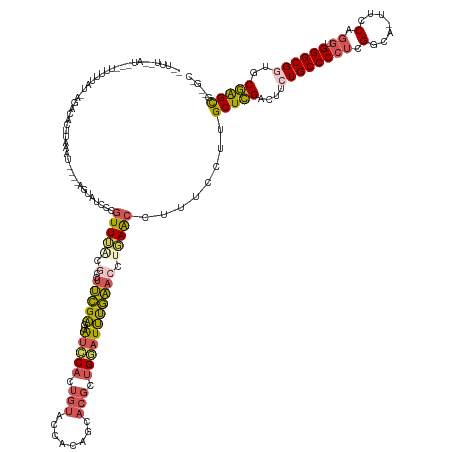
| Mean single sequence MFE | -40.96 |
| Consensus MFE | -23.38 |
| Energy contribution | -23.58 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.38 |
| Mean z-score | -0.78 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.766863 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
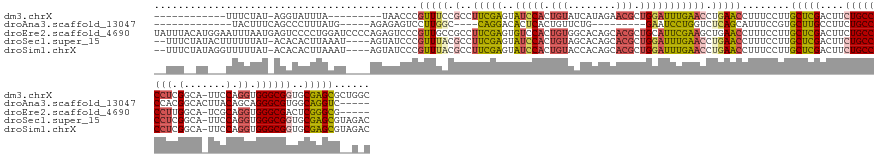
>dm3.chrX 9752990 133 - 22422827 ------------UUUCUAU-AGGUAUUUA---------UAACCCGUUUCCGCCUUCGAGUAUCCACUGUAUCAUAGAACGCUGGAUUUGAACCUGAACCUUUCCUUGCUCGACUUCUGCCCCUCGGCA-UUCCAGGUGGGCGGUGCGAGCGCUGGC ------------.......-.(((.....---------..))).......((((((((..(((((.(((........))).))))))))))...............(((((....(((((((..((..-..))..).))))))..)))))...))) ( -35.80, z-score = 0.02, R) >droAna3.scaffold_13047 324904 120 - 1816235 -------------UACUUUCAGCCCUUUAUG-----AGAGAGUCCUUGGC----CAGGACACUCACUGUUCUG---------GAAUCCUGGUCUCAGCAUUUCCGUGCUUGCCUUCUGCCCCACGGCACUUACAGCAGGGCGUGGCAGGUC----- -------------...(((((........))-----)))(((.((..(((----(((((((.....)))))))---------)...)).)).)))...........(((((((...(((((....((.......)).))))).))))))).----- ( -40.10, z-score = -0.48, R) >droEre2.scaffold_4690 13382719 150 + 18748788 UAUUUACAUGGAAUUUAAUGAGUCCCCUGGAUCCCCAGAGUCCCGUUGCCGCCUUCGAGUGUCCACUGUGGCACAGCACGCUGCAUUCGAAGCUGAACCUUUCCUUGCUCGACUUCUGCCCCUUGGCA-UCGCAGGUGGGCGACUCGGGCG----- .........((((..(((((.(..(.((((....)))).)..)))))).((.((((((((((....((((......))))..)))))))))).)).....))))..((((((..((.((((((((...-...)))).)))))).)))))).----- ( -48.90, z-score = -0.60, R) >droSec1.super_15 508230 148 - 1954846 --UUUCUAUACUUUUUUAU-ACACACUUAAAU----AGUAUCCCGUUUACGCCUUCGAGUAUCCACUGUAGCACAGCACGCUGGAUUUGAACCUGAACCUUUCCUUGCUCGACUUCUGCCCCUCGGCA-UUCCAGGUGGGCGGUGCGAGCGUAGAC --.....(((((..((((.-.......)))).----)))))...((((((...(((((..(((((.(((.((...))))).))))))))))...............(((((....(((((((..((..-..))..).))))))..))))))))))) ( -39.90, z-score = -1.61, R) >droSim1.chrX 7778118 148 - 17042790 --UUUCUAUAGGUUUUUAU-ACACACUUAAAU----AGUAUCCCGUUUACGCCUUCGAGUAUCCACUGUACCACAGCACGCUGGAUUUGAACCUGAACCUUUCCUUGCUCGACUUCUGCCCCUCGGCA-UUCCAGGUGGGCGGUGCGAGCGUAGAC --..(((((((((((....-....(((..((.----.(((.......)))...))..)))(((((.(((........))).)))))..))))))............(((((....(((((((..((..-..))..).))))))..)))))))))). ( -40.10, z-score = -1.21, R) >consensus __UUU__AU___UUUUUAU_AGACACUUAAAU____AGUAUCCCGUUUACGCCUUCGAGUAUCCACUGUACCACAGCACGCUGGAUUUGAACCUGAACCUUUCCUUGCUCGACUUCUGCCCCUCGGCA_UUCCAGGUGGGCGGUGCGAGCG__G_C ............................................(((((....(((((..(((((.(((........))).))))))))))..)))))........(((((....((((((((.(.......).)).))))))..)))))...... (-23.38 = -23.58 + 0.20)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:29:39 2011