| Sequence ID | dm3.chrX |
|---|---|
| Location | 9,747,218 – 9,747,363 |
| Length | 145 |
| Max. P | 0.628202 |

| Location | 9,747,218 – 9,747,363 |
|---|---|
| Length | 145 |
| Sequences | 12 |
| Columns | 149 |
| Reading direction | reverse |
| Mean pairwise identity | 89.04 |
| Shannon entropy | 0.24347 |
| G+C content | 0.42390 |
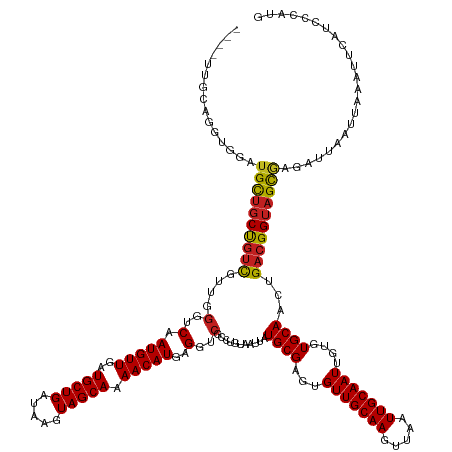
| Mean single sequence MFE | -42.48 |
| Consensus MFE | -35.74 |
| Energy contribution | -35.49 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.628202 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
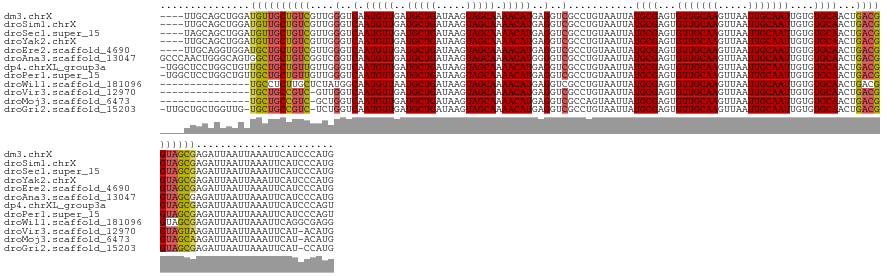
>dm3.chrX 9747218 145 - 22422827 ----UUGCAGCUGGAUGUUGCUGUCGUUGGGUCAAUGUUGAUGCUGAUAAGUAGCAAAACAUGAGGUCGCCUGUAAUUAUGCGAGUGUUGCAAGUUAAUUGCAAUUGUGUGCAACUGACGGUAGCGAGAUUAAUUAAAUUCAUCCCAUG ----........((((((((((..(((..(.(((.((((..(((((.....))))).)))))))...............((((...(((((((.....)))))))....)))).)..)))..)))))((((.....))))))))).... ( -39.90, z-score = -0.44, R) >droSim1.chrX 7772505 145 - 17042790 ----UUGCAGCUGGAUGUUGCUGUCGUUGGGUCAAUGUUGAUGCUGAUAAGUAGCAAAACAUGAGGUCGCCUGUAAUUAUGCGAGUGUUGCAAGUUAAUUGCAAUUGUGUGCAACUGACGGUAGCGAGAUUAAUUAAAUUCAUCCCAUG ----........((((((((((..(((..(.(((.((((..(((((.....))))).)))))))...............((((...(((((((.....)))))))....)))).)..)))..)))))((((.....))))))))).... ( -39.90, z-score = -0.44, R) >droSec1.super_15 502456 145 - 1954846 ----UAGCAGCUGGAUGUUGCUGUCGUUGGGUCAAUGUUGAUGCUGAUAAGUAGCAAAACAUGAGGUCGCCUGUAAUUAUGCGAGUGUUGCAAGUUAAUUGCAAUUGUGUGCAACUGACGGUAGCGAGAUUAAUUAAAUUCAUCCCAUG ----(((((((.....)))))))....(((((((.((((..(((((.....))))).)))))))..((((((((.....((((...(((((((.....)))))))....))))....))))..))))................)))).. ( -40.90, z-score = -0.76, R) >droYak2.chrX 18339082 145 - 21770863 ----UUGCAGCUGGAUGUUGCUGUCGUUGGGUCAAUGUUGAUGCUGAUAAGUAGCAAAACAUGAGGUCGCCUGUAAUUAUGCGAGUGUUGCAAGUUAAUUGCAAUUGUGUGCAACUGACGGUAGCGAGAUUAAUUAAAUUCAUCCCAUG ----........((((((((((..(((..(.(((.((((..(((((.....))))).)))))))...............((((...(((((((.....)))))))....)))).)..)))..)))))((((.....))))))))).... ( -39.90, z-score = -0.44, R) >droEre2.scaffold_4690 13377304 145 + 18748788 ----UUGCAGGUGGAUGCUGCUGUCGUUGGGUCAAUGUUGAUGCUGAUAAGUAGCAAAACAUGAGGUCGCCUGUAAUUAUGCGAGUGUUGCAAGUUAAUUGCAAUUGUGUGCAACUGACGGUAGCGAGAUUAAUUAAAUUCAUCCCAUG ----......((((.(((((((((((((((..(.(((((..(((((.....))))).)))))..)..)............(((...(((((((.....)))))))..))).)))).)))))))))).((..........))...)))). ( -42.70, z-score = -1.11, R) >droAna3.scaffold_13047 1480178 149 + 1816235 GCCCAACUGGGCAGUGGCUGCUGUCGGUCGGUCAAUGUUGAUGCUGAUAAGUAGCAAAACAUGGGGUCGCCUGUAAUUAUGCGAGUGUUGCAAGUUAAUUGCAAUUGUGUGCAACUGACGGUAGCGAGAUUAAUUAAAUUCAUCCCAUG ((((....))))....((((((((((((..((((....)))))))))).))))))....(((((((((((((((.....((((...(((((((.....)))))))....))))....))))..))))((..........)).))))))) ( -52.40, z-score = -2.58, R) >dp4.chrXL_group3a 977925 148 - 2690836 -UGGCUCCUGGCUGUUGCUGCUGUUGUUGGGUCAAUGUUGAUGCUGAUAAGUAGCAAAACAUGAGGUCGCCUGUAAUUAUGCGAGUGUUGCAAGUUAAUUGCAAUUGUGUGCAACUGACGGUAGCGAGAUUAAUUAAAUUCAUCCCAGU -......((((...((((((((((((((((..(.(((((..(((((.....))))).)))))..)..)............(((...(((((((.....)))))))..))).)))).)))))))))))((..........))...)))). ( -42.10, z-score = -0.51, R) >droPer1.super_15 261971 148 + 2181545 -UGGCUCCUGGCUGUUGCUGCUGUUGUUGGGUCAAUGUUGAUGCUGAUAAGUAGCAAAACAUGAGGUCGCCUGUAAUUAUGCGAGUGUUGCAAGUUAAUUGCAAUUGUGUGCAACUGACGGUAGCGAGAUUAAUUAAAUUCAUCCCAGU -......((((...((((((((((((((((..(.(((((..(((((.....))))).)))))..)..)............(((...(((((((.....)))))))..))).)))).)))))))))))((..........))...)))). ( -42.10, z-score = -0.51, R) >droWil1.scaffold_181096 9239673 134 + 12416693 ---------------UGCCUCUUGCUCUAUGGCAAUGUUAAUGCUGAUAAGUAGCAAAACAUGAGGUCGCCUGUAAUUAUGCGAGUGUUGCAAGUUAAUUGCAAUUGUGUGCAACUGACGGUAGCGAGAUUAAUUAAAUUCAGGCGAGG ---------------..((((.((((....))))(((((..(((((.....))))).)))))))))(((((((.((((.....(((.((((..((((.(((((......))))).))))....)))).))).....))))))))))).. ( -41.60, z-score = -2.05, R) >droVir3.scaffold_12970 2835852 132 + 11907090 ---------------UGCUGCCGUC-GUUGGUCAAUGUUGAUGCUGAUAAGUAGCAAAACAUGAGGUCGCCUGUAAUUAUGCGAGUGUUGCAAGUUAAUUGCAAUUGUGUGCAACUGACGGUAGUAAGAUUAAUUAAAUUCAU-ACAUG ---------------((((((((((-(.((..(.(((((..(((((.....))))).)))))..)..)).)........((((...(((((((.....)))))))....))))...)))))))))).................-..... ( -39.50, z-score = -2.15, R) >droMoj3.scaffold_6473 16394467 132 - 16943266 ---------------UGCUGCCGUC-GCUGGUCAAUGUUGAUGCUGAUAAGUAGCAAAACAUGAGGUCGCCAGUAAUUAUGCGAGUGUUGCAAGUUAAUUGCAAUUGUGUGCAACUGACGGUAGCAAGAUUAAUUAAAUUCAU-ACAUG ---------------((((((((((-(((((((.(((((..(((((.....))))).)))))...)..)))))).....((((...(((((((.....)))))))....))))...)))))))))).................-..... ( -45.90, z-score = -3.53, R) >droGri2.scaffold_15203 248481 145 + 11997470 -UUGCUGCUGGUUG-UGCUGCCGUC-UCUGGUCAAUGUUGAUGCUGAUAAGUAGCAAAACAUGAGGUCGCCUGUAAUUAUGCGAGUGUUGCAAGUUAAUUGCAAUUGUGUGCAACUGACGGUAGCGAGAUUAAUUAAAUUCAU-CCAUG -.......(((((.-((((((((((-...(..(.(((((..(((((.....))))).)))))..)..)...........((((...(((((((.....)))))))....))))...)))))))))).)))))...........-..... ( -42.80, z-score = -1.25, R) >consensus ____UUGCAGGUGGAUGCUGCUGUCGUUGGGUCAAUGUUGAUGCUGAUAAGUAGCAAAACAUGAGGUCGCCUGUAAUUAUGCGAGUGUUGCAAGUUAAUUGCAAUUGUGUGCAACUGACGGUAGCGAGAUUAAUUAAAUUCAUCCCAUG ...............((((((((((....(..(.(((((..(((((.....))))).)))))..)..)...........((((...(((((((.....)))))))....))))...))))))))))....................... (-35.74 = -35.49 + -0.25)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:29:38 2011