
| Sequence ID | dm3.chrX |
|---|---|
| Location | 9,744,369 – 9,744,462 |
| Length | 93 |
| Max. P | 0.643073 |

| Location | 9,744,369 – 9,744,462 |
|---|---|
| Length | 93 |
| Sequences | 12 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 80.31 |
| Shannon entropy | 0.41015 |
| G+C content | 0.38386 |
| Mean single sequence MFE | -21.00 |
| Consensus MFE | -13.21 |
| Energy contribution | -12.99 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.643073 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 9744369 93 - 22422827 UAUUUAAACGUGAUUUUAAAUAACCGCAACAGAUUUUGUGGUUUCAUGUUGCAUUUGCCGUUCGAUAAGCGACAGCCCGACAUUGCCGGGUGG---- ((((((((......))))))))(((((((......)))))))....((((((..(((.....)))...))))))(((((.......)))))..---- ( -23.40, z-score = -0.85, R) >droSim1.chrX 7769428 93 - 17042790 UAUUUAAACGUGAUUUUAAAUAACCGCAACAGAUUUUGUGGUUUCAUGUUGCAUUUGCCGUUCGAUAAGCGACAGCCCGACAUUCCCGGGUGG---- ((((((((......))))))))(((((((......)))))))....((((((..(((.....)))...))))))(((((.......)))))..---- ( -23.40, z-score = -1.18, R) >droSec1.super_15 499630 93 - 1954846 UAUUUAAACGUGAUUUUAAAUAACCGCAACAGAUUUUGUGGAUUCAUGUUGCAUUUGCCGUUCGAUAAGCGACAGCCCGACAUUCCCGGGUGG---- ((((((((......)))))))).((((((......)))))).....((((((..(((.....)))...))))))(((((.......)))))..---- ( -22.90, z-score = -1.09, R) >droYak2.chrX 18336064 93 - 21770863 CAUUUAAACGUGAUUUUAAAUAACCGCAACAGAUUUUGUGGUUUCAUGUUGCAUUUGCCGUUCGAUAAGCGACAGCCCGACAUUCCCGGGUGG---- ......(((((((........((((((((......))))))))))))))).....((.((((.....)))).))(((((.......)))))..---- ( -23.20, z-score = -1.03, R) >droEre2.scaffold_4690 13374540 93 + 18748788 UAUUUAAACGUGAUUUUAAAUAACCGCAACAGAUUUUGUGGUUUCAUGUUGCAUUUGCCGUUCGAUAAGCGACAGCCCGACAUUCCCGGGUGG---- ((((((((......))))))))(((((((......)))))))....((((((..(((.....)))...))))))(((((.......)))))..---- ( -23.40, z-score = -1.18, R) >droAna3.scaffold_13047 1477789 92 + 1816235 UAUUUAAACGUGAUUUUAAAUAACCACAACAGAUUUUGUGGUUUCAUGUUGCAUUUG-CGUUCGAUAAGGGACAGCCUGACGGUCCCGGGUGG---- ......(((((((........((((((((......)))))))))))))))((....)-)...(.((..(((((.(.....).)))))..)).)---- ( -23.80, z-score = -0.99, R) >dp4.chrXL_group3a 974891 94 - 2690836 UAUUUAAACGUGAUUUUAAAUAACCGCAACAGAUUUUGUGGUUUCAUGUUGCAUUUGCUGUUCGAUAAGCGACAG---GACAGCCUCUGGGCUCUGG ......(((((((........((((((((......)))))))))))))))....(((((........)))))(((---...((((....))))))). ( -24.80, z-score = -1.51, R) >droPer1.super_15 258934 94 + 2181545 UAUUUAAACGUGAUUUUAAAUAACCGCAACAGAUUUUGUGGUUUCAUGUUGCAUUUGCUGUUCGAUAAGCGACAG---GACAGCCUCUGGGCUCUGG ......(((((((........((((((((......)))))))))))))))....(((((........)))))(((---...((((....))))))). ( -24.80, z-score = -1.51, R) >droWil1.scaffold_181096 9235801 87 + 12416693 UAUUUAAACGUGAUUUUAAAUAACCGCAACAGAUUUUGUGGUUUCAUGUUGCAUUU--CGCUGUGUGUUCGAUAA-GGGACAUUGUAUGG------- ......(((((((........((((((((......)))))))))))))))......--.((...((((((.....-.)))))).))....------- ( -19.00, z-score = -0.78, R) >droVir3.scaffold_12970 2832805 71 + 11907090 UAUUUAAACGUGAUUUAAAAUAACCACAACAGAUUUUGUGGUUUUAUGUUGCAUUUUAUGUUUGAUAAUCG-------------------------- ...((((((((((.....(((((((((((......)))))))....)))).....))))))))))......-------------------------- ( -15.90, z-score = -2.73, R) >droMoj3.scaffold_6473 16391027 71 - 16943266 UAUUUAAACGUGAUUUAAAAUAACCACAACAGAUUUUGUGGUUUCAUGUUGCAUUUUAUGUUCGAUAAGCG-------------------------- ......(((((((........((((((((......))))))))))))))).....................-------------------------- ( -13.70, z-score = -1.52, R) >droGri2.scaffold_15203 245492 70 + 11997470 -AUUUAAACGUGAUUUUAAAUAACCACAACAGAUUUUGUGGUUUCAUGUUGCAUUUUAUAUUCGAUAAGCG-------------------------- -.....(((((((........((((((((......))))))))))))))).....................-------------------------- ( -13.70, z-score = -2.06, R) >consensus UAUUUAAACGUGAUUUUAAAUAACCGCAACAGAUUUUGUGGUUUCAUGUUGCAUUUGCCGUUCGAUAAGCGACAG_C_GACAUUCCCGGGUGG____ ......(((((((........((((((((......)))))))))))))))............................................... (-13.21 = -12.99 + -0.21)

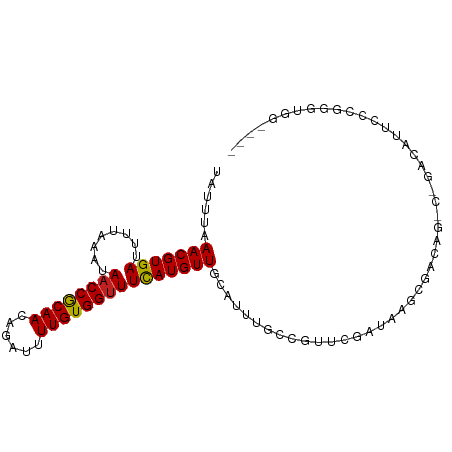
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:29:37 2011