| Sequence ID | dm3.chrX |
|---|---|
| Location | 9,709,301 – 9,709,398 |
| Length | 97 |
| Max. P | 0.928123 |

| Location | 9,709,301 – 9,709,398 |
|---|---|
| Length | 97 |
| Sequences | 8 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 65.42 |
| Shannon entropy | 0.68240 |
| G+C content | 0.54634 |
| Mean single sequence MFE | -18.37 |
| Consensus MFE | -5.40 |
| Energy contribution | -5.34 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.29 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.928123 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
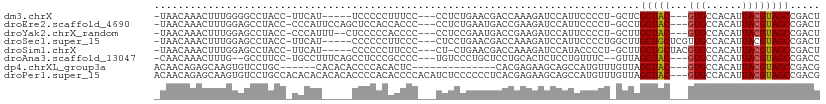
>dm3.chrX 9709301 97 - 22422827 -UAACAAACUUUGGGGCCUACC-UUCAU-----UCCCCCUUUCC---CCUCUGAACGACCAAAGAUCCAUUCCCCU-GCUCGCUAC---GUGCCACAUUACGUAGCCGACU -.......((((((((....))-.....-----.....(.(((.---.....))).).))))))............-....(((((---(((......))))))))..... ( -17.60, z-score = -1.86, R) >droEre2.scaffold_4690 13338820 102 + 18748788 -UAACAAACUUUGGAGCCUACC-CCCAUUCCAGCUCCACCACCC---CCUCUGAAUGACCGAAGAUCCAUUCCCCU-GCCUGCUAC---GUGCCACAUUACGUAGCCGACU -..........((((((.....-.........))))))......---.....(((((..........)))))....-..(.(((((---(((......)))))))).)... ( -20.34, z-score = -3.77, R) >droYak2.chrX_random 229439 100 - 1802292 -UAACAAACUUUGGAGCCUACC-CCCAUUU--CUCCCCCACCCC---CCUCCGAAUGACCGAAGAUCCAUUCCCCU-GCUUGCUAC---GUGCCACAUUACGUAGCCGACU -...........((((......-.......--............---.))))(((((..........)))))....-....(((((---(((......))))))))..... ( -15.47, z-score = -2.67, R) >droSec1.super_15 465728 101 - 1954846 -UAACAAACUUUGGAGCCUACC-UUCAU-----CCCCCCUUCCC---CUCCUGAACGACCAAAGAUCCAUUCCCCUGGCUUGCUGCUCGUUGCCACAUUACGUAGCCGACU -..........(((((.....)-)))).-----...........---......(((((.((..((.(((......))).))..)).)))))((.((.....)).))..... ( -12.00, z-score = 0.39, R) >droSim1.chrX 7740428 99 - 17042790 -UAACAAACUUUGGAGCCUACC-UUCAU-----CCCCCCUUCCC---CU-CUGAACGACCAAAGAUCCAUACCCCU-GCUUGCUGCUACGUGCCACAUUACGUAGCCGACU -.......((((((........-.....-----...........---..-........))))))............-.....(.((((((((......)))))))).)... ( -14.48, z-score = -1.86, R) >droAna3.scaffold_13047 1445761 99 + 1816235 -CAACAAACUUUG--GCCUUCC-UGCCUUUCAGCCUCCCGCCCC---UGUCCCUGCUCCUGCACUCUCCUGUUUC--GUUAGCUAC---GUGCCACAUUACGUAGCCGACC -.((((......(--((.....-.((......)).....)))..---......(((....)))......))))..--((..(((((---(((......))))))))..)). ( -19.90, z-score = -2.27, R) >dp4.chrXL_group3a 940263 88 - 2690836 ACAACAGAGCAAGUGUCCUGC------CACACACCCCACACUC--------------CACGAGAAGCAGCCAUGUUUGUUAGCUAC---GUGCCACAUUACGUAGCCGACG ..((((..((..((((.....------.))))........(((--------------...))).....))..)))).((..(((((---(((......))))))))..)). ( -22.80, z-score = -2.51, R) >droPer1.super_15 222949 108 + 2181545 ACAACAGAGCAAGUGUCCUGCCACACACACACACCCCACACCCCACAUCUCCCCCCUCACGAGAAGCAGCCAUGUUUGUUAGCUAC---GUGCCACAUUACGUAGCCGACG ..((((..((..((((......)))).....................((((.........))))....))..)))).((..(((((---(((......))))))))..)). ( -24.40, z-score = -3.42, R) >consensus _UAACAAACUUUGGAGCCUACC_UUCAU_____CCCCCCACCCC___CCUCUGAACGACCGAAGAUCCAUUCCCCU_GCUUGCUAC___GUGCCACAUUACGUAGCCGACU .................................................................................(((((...(((...)))...)))))..... ( -5.40 = -5.34 + -0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:29:31 2011