
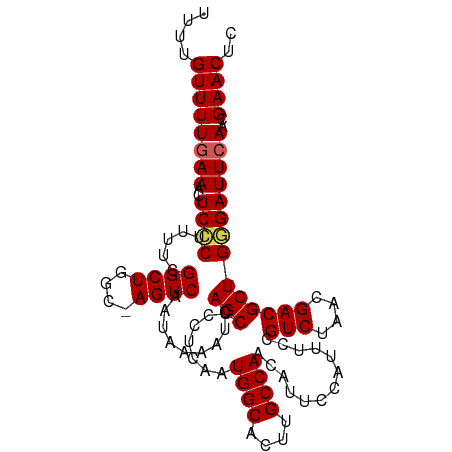
| Sequence ID | dm3.chrX |
|---|---|
| Location | 9,688,137 – 9,688,282 |
| Length | 145 |
| Max. P | 0.648415 |

| Location | 9,688,137 – 9,688,244 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 95.25 |
| Shannon entropy | 0.08415 |
| G+C content | 0.44775 |
| Mean single sequence MFE | -23.35 |
| Consensus MFE | -20.02 |
| Energy contribution | -22.18 |
| Covariance contribution | 2.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.556428 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 9688137 107 + 22422827 UUUUGUUUUGAAACUCCCUUUUCGGCUGGC-AGUCAAUAAAAUCAGCUCCUCAAUGGCACUUGCCAACAUUCCAUUUCCGUCUAACGACGCUGUGAUUCAACGAACUC .(((((..((((...........(((....-.)))........((((.......((((....)))).............(((....)))))))...)))))))))... ( -20.90, z-score = -1.08, R) >droEre2.scaffold_4690 13325464 108 - 18748788 CAUUGUUUUAAAACUCCCUUUUCGGCUGGCCAGUCAAUAAAAUCAGCUCUUCAAUGGCACUUGCCAACAUUCCAUUUCCGUCUAACGACGCUGGGAUUCAACGAACCC ....((((......((((.....((((....)))).........(((.......((((....)))).............(((....))))))))))......)))).. ( -23.30, z-score = -1.59, R) >droYak2.chrX 18291243 107 + 21770863 CUCUGUUUUGAAACUCGCUUUUCGGCUGCC-AGUCAAUAAAAUCAGCUCUUCAAUGGCACUUGCCAACAUUCCAUUUCCGUCUAACGACGCUGCGAUUCAACGAACUC ....((((((((..((((.....(((((((-(......................)))))...))).............((((....))))..))))))))..)))).. ( -23.35, z-score = -2.03, R) >droSec1.super_15 451720 107 + 1954846 UUUUGUUUUGAAACUCCCUUUUCGGCUGGC-AGUCAAUAAAAUCAGCUCCUCAAUGGCACUUGCCAACAUUCCAUUUCCGUCUAACGACGCUGGGAUUCAACGAACUC .(((((..((((..((((.....((((((.-...........))))))......((((....))))............((((....))))..)))))))))))))... ( -24.60, z-score = -1.77, R) >droSim1.chrU 294649 107 - 15797150 UUUUGUUUUGAAACUCCCUUUUCGGCUGGC-AGUCAAUAAAAUCAGCUUCUCAAUGGCACUUGCCAACAUUCCAUUUCCGUCUAACGACGCUGGGAUUCAACGAACUC .(((((..((((..((((.....((((((.-...........))))))......((((....))))............((((....))))..)))))))))))))... ( -24.60, z-score = -1.85, R) >consensus UUUUGUUUUGAAACUCCCUUUUCGGCUGGC_AGUCAAUAAAAUCAGCUCCUCAAUGGCACUUGCCAACAUUCCAUUUCCGUCUAACGACGCUGGGAUUCAACGAACUC ....((((((((..((((.....((((....)))).........(((.......((((....)))).............(((....))))))))))))))..)))).. (-20.02 = -22.18 + 2.16)

| Location | 9,688,175 – 9,688,282 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 76.18 |
| Shannon entropy | 0.45689 |
| G+C content | 0.46958 |
| Mean single sequence MFE | -22.91 |
| Consensus MFE | -11.43 |
| Energy contribution | -11.55 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.648415 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
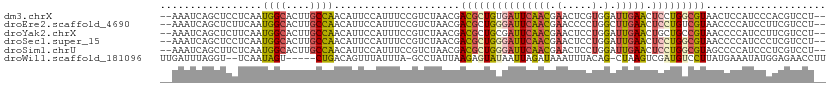
>dm3.chrX 9688175 107 + 22422827 --AAAUCAGCUCCUCAAUGGCACUUGCCAACAUUCCAUUUCCGUCUAACGACGCUGUGAUUCAACGAACUCGUGGAUUGAACUCCUGGCGUAACUCCAUCCCACGUCCU-- --....((((.......((((....)))).............(((....))))))).((((((.((....))))))))........(((((...........)))))..-- ( -23.30, z-score = -1.71, R) >droEre2.scaffold_4690 13325503 107 - 18748788 --AAAUCAGCUCUUCAAUGGCACUUGCCAACAUUCCAUUUCCGUCUAACGACGCUGGGAUUCAACGAACCCCUGGCUUGAACUCCUGUCGUAACCCCAUCCUUCGUCCU-- --....(((...(((((((((....)))).............(((....)))((((((.(((...))).))).))))))))...)))......................-- ( -21.40, z-score = -1.26, R) >droYak2.chrX 18291281 107 + 21770863 --AAAUCAGCUCUUCAAUGGCACUUGCCAACAUUCCAUUUCCGUCUAACGACGCUGCGAUUCAACGAACUCCUGGAUUGAACUGCUGCCGUAACCCCAUCCUUCGUCCU-- --....((((..(((((((((....))))....((((....((((....))))...((......))......)))))))))..))))......................-- ( -23.50, z-score = -2.67, R) >droSec1.super_15 451758 107 + 1954846 --AAAUCAGCUCCUCAAUGGCACUUGCCAACAUUCCAUUUCCGUCUAACGACGCUGGGAUUCAACGAACUCCUGGAUUGAACUCCUGGCGUAACCCCAUCCCUCGUCCU-- --.....(((.......((((....)))).............(((....))))))(((((.......((.((.(((......))).)).))......))))).......-- ( -25.22, z-score = -2.00, R) >droSim1.chrU 294687 107 - 15797150 --AAAUCAGCUUCUCAAUGGCACUUGCCAACAUUCCAUUUCCGUCUAACGACGCUGGGAUUCAACGAACUCCUGGAUUGAACUCCUGGCGUAGCCCCAUCCCUCGUCCU-- --.....(((.......((((....)))).............(((....))))))(((((.......((.((.(((......))).)).))......))))).......-- ( -25.22, z-score = -1.41, R) >droWil1.scaffold_181096 9180539 102 - 12416693 UUGAUUUAGGU--UCAAUAGU-----CUGACAGUUUAUUUA-GCCUAUUAAGAGUAUAAUUAGAUAAAUUUACAG-CUAAGUCGAUGUCCUUAUGAAAUAUGGAGAACCUU .......((((--((..(((.-----(((..((((((((((-(..(((.......))).)))))))))))..)))-)))........(((...........))))))))). ( -18.80, z-score = -0.80, R) >consensus __AAAUCAGCUCCUCAAUGGCACUUGCCAACAUUCCAUUUCCGUCUAACGACGCUGGGAUUCAACGAACUCCUGGAUUGAACUCCUGGCGUAACCCCAUCCCUCGUCCU__ .................((((....)))).....................((((((((((((((....(....)..))))).))))))))).................... (-11.43 = -11.55 + 0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:29:28 2011