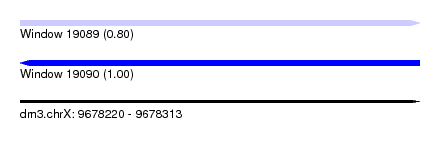
| Sequence ID | dm3.chrX |
|---|---|
| Location | 9,678,220 – 9,678,313 |
| Length | 93 |
| Max. P | 0.996566 |

| Location | 9,678,220 – 9,678,313 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 78.61 |
| Shannon entropy | 0.39951 |
| G+C content | 0.49679 |
| Mean single sequence MFE | -25.62 |
| Consensus MFE | -16.08 |
| Energy contribution | -16.25 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.799824 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
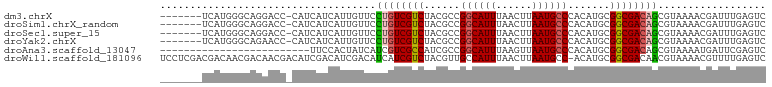
>dm3.chrX 9678220 93 + 22422827 -------UCAUGGGCAGGACC-CAUCAUCAUUGUUCCUGUCGUCUACGCCGGCAUUUAACUUAAUGCCCACAUGCGGCGACAGCGUAAAACGAUUUGAGUC -------...((((.....))-))(((..(((((((((((((((((.(..((((((......))))))..).)).)))))))).)...)))))).)))... ( -27.50, z-score = -1.61, R) >droSim1.chrX_random 2774310 93 + 5698898 -------UCAUGGGCAGGACC-CAUCAUCAUUGUUCCUGUCGUCUACGCCGGCAUUUAACUUAAUGCCCACAUGCGGCGACAGCGUAAAACGAUUUGAGUC -------...((((.....))-))(((..(((((((((((((((((.(..((((((......))))))..).)).)))))))).)...)))))).)))... ( -27.50, z-score = -1.61, R) >droSec1.super_15 441991 93 + 1954846 -------UCAUGGGCAGGACC-CAUCAUCAUUGUUCCUGUCGUCUACGCCGGCAUUUAACUUAAUGCCCACAUGCGGCGACAGCGUAAAACGAUUUGAGUC -------...((((.....))-))(((..(((((((((((((((((.(..((((((......))))))..).)).)))))))).)...)))))).)))... ( -27.50, z-score = -1.61, R) >droYak2.chrX 18279802 93 + 21770863 -------UCAUGGGCAGAACC-CAUCAUCAUUGUUCCUGUCGUCUACGCCGGCAUUUAACUUAAUGCCCACAUGCGGCGACAGCGUAAAACGAUUUGAGUC -------...((((.....))-))(((..(((((((((((((((((.(..((((((......))))))..).)).)))))))).)...)))))).)))... ( -27.50, z-score = -2.08, R) >droAna3.scaffold_13047 1426714 76 - 1816235 -------------------------UUCCACUAUCAUCGUCGCCAUCGCCGGCAUUUAAGUUAAUGCCCACAUGCGGCGACAGCGUAAAAUGAUUCGAGUC -------------------------(((....(((((..((((..(((((((((((......)))))).......)))))..))).)..)))))..))).. ( -21.51, z-score = -2.09, R) >droWil1.scaffold_181096 9164960 100 - 12416693 UCCUCGACGACAACGACAACGACAUCGACAUCGACAUCAUCGUCUACGUUGCCAUUUAACUUAAUGCC-ACAUGCGGCGACAACGUAAAACGUUUUGAGUC ..(((((.(((...(((...((..(((....)))..))...)))(((((((.((((......)))(((-......)))).)))))))....)))))))).. ( -22.20, z-score = -0.78, R) >consensus _______UCAUGGGCAGGACC_CAUCAUCAUUGUUCCUGUCGUCUACGCCGGCAUUUAACUUAAUGCCCACAUGCGGCGACAGCGUAAAACGAUUUGAGUC ....................................((((((((......((((((......)))))).......)))))))).................. (-16.08 = -16.25 + 0.17)

| Location | 9,678,220 – 9,678,313 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 78.61 |
| Shannon entropy | 0.39951 |
| G+C content | 0.49679 |
| Mean single sequence MFE | -33.95 |
| Consensus MFE | -22.79 |
| Energy contribution | -22.77 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.37 |
| Mean z-score | -2.84 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.95 |
| SVM RNA-class probability | 0.996566 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
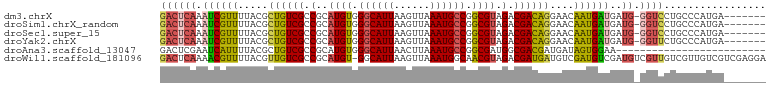
>dm3.chrX 9678220 93 - 22422827 GACUCAAAUCGUUUUACGCUGUCGCCGCAUGUGGGCAUUAAGUUAAAUGCCGGCGUAGACGACAGGAACAAUGAUGAUG-GGUCCUGCCCAUGA------- ((((((.((((((...(.((((((.(..((((.((((((......)))))).)))).).)))))))...))))))..))-))))..........------- ( -34.20, z-score = -2.70, R) >droSim1.chrX_random 2774310 93 - 5698898 GACUCAAAUCGUUUUACGCUGUCGCCGCAUGUGGGCAUUAAGUUAAAUGCCGGCGUAGACGACAGGAACAAUGAUGAUG-GGUCCUGCCCAUGA------- ((((((.((((((...(.((((((.(..((((.((((((......)))))).)))).).)))))))...))))))..))-))))..........------- ( -34.20, z-score = -2.70, R) >droSec1.super_15 441991 93 - 1954846 GACUCAAAUCGUUUUACGCUGUCGCCGCAUGUGGGCAUUAAGUUAAAUGCCGGCGUAGACGACAGGAACAAUGAUGAUG-GGUCCUGCCCAUGA------- ((((((.((((((...(.((((((.(..((((.((((((......)))))).)))).).)))))))...))))))..))-))))..........------- ( -34.20, z-score = -2.70, R) >droYak2.chrX 18279802 93 - 21770863 GACUCAAAUCGUUUUACGCUGUCGCCGCAUGUGGGCAUUAAGUUAAAUGCCGGCGUAGACGACAGGAACAAUGAUGAUG-GGUUCUGCCCAUGA------- .......((((((...(.((((((.(..((((.((((((......)))))).)))).).)))))))...)))))).(((-((.....)))))..------- ( -33.10, z-score = -2.46, R) >droAna3.scaffold_13047 1426714 76 + 1816235 GACUCGAAUCAUUUUACGCUGUCGCCGCAUGUGGGCAUUAACUUAAAUGCCGGCGAUGGCGACGAUGAUAGUGGAA------------------------- .(((...((((((...((((((((((((((.((((......)))).))).)))))))))))..)))))))))....------------------------- ( -29.00, z-score = -3.25, R) >droWil1.scaffold_181096 9164960 100 + 12416693 GACUCAAAACGUUUUACGUUGUCGCCGCAUGU-GGCAUUAAGUUAAAUGGCAACGUAGACGAUGAUGUCGAUGUCGAUGUCGUUGUCGUUGUCGUCGAGGA (((....(((....(((((((((((((....)-)))(((......))))))))))))(((((((((((((....))))))))))))))))...)))..... ( -39.00, z-score = -3.25, R) >consensus GACUCAAAUCGUUUUACGCUGUCGCCGCAUGUGGGCAUUAAGUUAAAUGCCGGCGUAGACGACAGGAACAAUGAUGAUG_GGUCCUGCCCAUGA_______ ((((...((((((.....((((((.(..((((.((((((......)))))).)))).).))))))....)))))).....))))................. (-22.79 = -22.77 + -0.02)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:29:26 2011