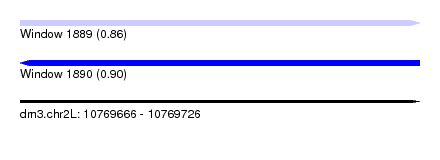
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 10,769,666 – 10,769,726 |
| Length | 60 |
| Max. P | 0.901212 |

| Location | 10,769,666 – 10,769,726 |
|---|---|
| Length | 60 |
| Sequences | 8 |
| Columns | 60 |
| Reading direction | forward |
| Mean pairwise identity | 76.85 |
| Shannon entropy | 0.49428 |
| G+C content | 0.54627 |
| Mean single sequence MFE | -12.80 |
| Consensus MFE | -8.41 |
| Energy contribution | -9.10 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.863253 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
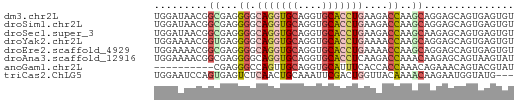
>dm3.chr2L 10769666 60 + 23011544 ACACUCACUGCUCCUGCUUGGUCUUCAGGUGCACCUGCACCUGCCCCUCGCCGUUAUCCA ...............((..((....(((((((....)))))))..))..))......... ( -15.40, z-score = -2.02, R) >droSim1.chr2L 10570705 60 + 22036055 ACACUCACUGCUCCUGCUUGGUCUUCAGGUGCACCUGCACCUGCCCCUCGCCGUUAUCCA ...............((..((....(((((((....)))))))..))..))......... ( -15.40, z-score = -2.02, R) >droSec1.super_3 6188128 60 + 7220098 ACACUCACUGCUCUUGCUUGGUCUUCAGGUGCACCUGCACCUGCCCCUCGCCGUUAUCCA ...............((..((....(((((((....)))))))..))..))......... ( -15.40, z-score = -2.07, R) >droYak2.chr2L 7175040 60 + 22324452 ACACUCACUGCUCCUGCUUGGUUUUCAGGUGCACCUGCACCUGCCCCUCACCGUUUUCCA .........((....))..(((...(((((((....)))))))......)))........ ( -15.00, z-score = -2.31, R) >droEre2.scaffold_4929 11979889 60 - 26641161 ACACUCACUGCUCCUGCUUGGUUUUCAGGUGCACCUGCACCUGCCCCUCGCCGUUUUCCA ...............((..((....(((((((....)))))))..))..))......... ( -15.40, z-score = -2.11, R) >droAna3.scaffold_12916 14620265 60 + 16180835 AUACUUACUGCUCUUGUUUGGUCUUGAGGUGCACCUGCACCUGCCCCUCGCCGUUUUCCA ...................(((...(((((((....)))))).).....)))........ ( -11.40, z-score = -0.67, R) >anoGam1.chr2L 43949677 50 + 48795086 AUACGUACUGUUUCUGUUUGGUGGUGAAAUGCACCUGCAACUGGCCCUCG---------- ...................((.(((.(..(((....)))..).))).)).---------- ( -7.40, z-score = 1.02, R) >triCas2.ChLG5 10249478 57 + 18847211 ---CAUACCAUUCUUGUUUUGUAACCAGUCGAAUUUGCAGUUGAGACUCACUGGAUUCCA ---.....................(((((.((.(((.......))).)))))))...... ( -7.00, z-score = 0.76, R) >consensus ACACUCACUGCUCCUGCUUGGUCUUCAGGUGCACCUGCACCUGCCCCUCGCCGUUUUCCA ...............((..((....(((((((....)))))))..))..))......... ( -8.41 = -9.10 + 0.69)
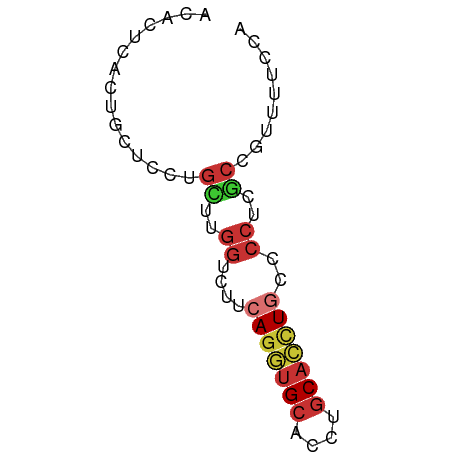
| Location | 10,769,666 – 10,769,726 |
|---|---|
| Length | 60 |
| Sequences | 8 |
| Columns | 60 |
| Reading direction | reverse |
| Mean pairwise identity | 76.85 |
| Shannon entropy | 0.49428 |
| G+C content | 0.54627 |
| Mean single sequence MFE | -15.01 |
| Consensus MFE | -11.60 |
| Energy contribution | -12.23 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.01 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.901212 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 10769666 60 - 23011544 UGGAUAACGGCGAGGGGCAGGUGCAGGUGCACCUGAAGACCAAGCAGGAGCAGUGAGUGU .........((..((..(((((((....)))))))....))..))............... ( -18.70, z-score = -1.73, R) >droSim1.chr2L 10570705 60 - 22036055 UGGAUAACGGCGAGGGGCAGGUGCAGGUGCACCUGAAGACCAAGCAGGAGCAGUGAGUGU .........((..((..(((((((....)))))))....))..))............... ( -18.70, z-score = -1.73, R) >droSec1.super_3 6188128 60 - 7220098 UGGAUAACGGCGAGGGGCAGGUGCAGGUGCACCUGAAGACCAAGCAAGAGCAGUGAGUGU .........((..((..(((((((....)))))))....))..))............... ( -18.70, z-score = -1.91, R) >droYak2.chr2L 7175040 60 - 22324452 UGGAAAACGGUGAGGGGCAGGUGCAGGUGCACCUGAAAACCAAGCAGGAGCAGUGAGUGU ........(((......(((((((....)))))))...)))..((....))......... ( -17.70, z-score = -1.68, R) >droEre2.scaffold_4929 11979889 60 + 26641161 UGGAAAACGGCGAGGGGCAGGUGCAGGUGCACCUGAAAACCAAGCAGGAGCAGUGAGUGU .........((..((..(((((((....)))))))....))..))............... ( -18.70, z-score = -1.76, R) >droAna3.scaffold_12916 14620265 60 - 16180835 UGGAAAACGGCGAGGGGCAGGUGCAGGUGCACCUCAAGACCAAACAAGAGCAGUAAGUAU (((......((.....))((((((....)))))).....))).................. ( -13.00, z-score = -0.49, R) >anoGam1.chr2L 43949677 50 - 48795086 ----------CGAGGGCCAGUUGCAGGUGCAUUUCACCACCAAACAGAAACAGUACGUAU ----------...((((.....)).((((.....)))).))................... ( -7.10, z-score = 0.54, R) >triCas2.ChLG5 10249478 57 - 18847211 UGGAAUCCAGUGAGUCUCAACUGCAAAUUCGACUGGUUACAAAACAAGAAUGGUAUG--- ((....(((((((((...........)))).)))))...))................--- ( -7.50, z-score = 0.67, R) >consensus UGGAAAACGGCGAGGGGCAGGUGCAGGUGCACCUGAAGACCAAGCAGGAGCAGUGAGUGU .........((...((.(((((((....)))))))....))..))............... (-11.60 = -12.23 + 0.63)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:30:57 2011