
| Sequence ID | dm3.chrX |
|---|---|
| Location | 9,653,270 – 9,653,361 |
| Length | 91 |
| Max. P | 0.869811 |

| Location | 9,653,270 – 9,653,361 |
|---|---|
| Length | 91 |
| Sequences | 7 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 72.74 |
| Shannon entropy | 0.46318 |
| G+C content | 0.49434 |
| Mean single sequence MFE | -29.80 |
| Consensus MFE | -21.55 |
| Energy contribution | -21.59 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.869811 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
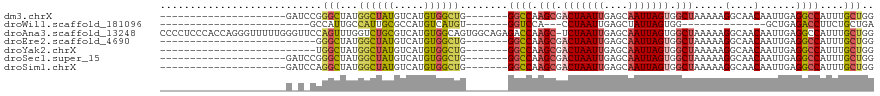
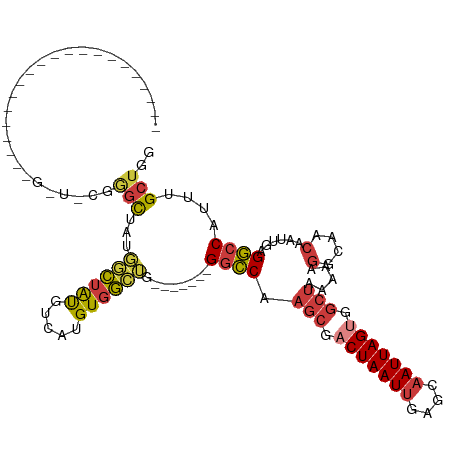
>dm3.chrX 9653270 91 - 22422827 ---------------------GAUCCGGGCUAUGGCUAUGUCAUGUGGCUG-------GGCCAAGCGACUAAUUGAGCAAUUAGUGGCUAAAAAGGCAACAAUUGAGGCCAUUUGCUGG ---------------------...((((.....((((((.....)))))).-------((((.(((.(((((((....))))))).))).....(....)......)))).....)))) ( -30.60, z-score = -1.46, R) >droWil1.scaffold_181096 9124734 70 + 12416693 -------------------------GCCAUUGCCAUUGCGCCAUGUCAUGU-------GGUCCA---CCUAAUUGAGCUAUUAGUGG--------------GCUGAGACCUUCUGCUGA -------------------------............(((....(((...(-------((((((---(.((((......))))))))--------------)))).)))....)))... ( -15.70, z-score = 0.87, R) >droAna3.scaffold_13248 1490601 118 - 4840945 CCCCUCCCACCAGGGUUUUUGGGUUCCAGUUUGGUCUGCGUCAUGUGGCAGUGGCAGAGACCAAGC-UCUAAUUGAGCAAUUAGUGGCUAAAAAGGCAACAAUUGAGGCCAUUUGCUGG .(((.(((....))).....)))..((((((((((((..(((((......)))))..)))))))((-((.....))))....(((((((.((..(....)..))..))))))).))))) ( -41.60, z-score = -1.22, R) >droEre2.scaffold_4690 13288038 86 + 18748788 --------------------------GGGCUAUGGCUAUGUCAUGUGGCUG-------GGCCAAGCGACUAAUUGAGCAAUUAGUGGCUAAAAAGGCAACAAUUGAGGCCAUUUGCUGG --------------------------.(((...((((((.....)))))).-------((((.(((.(((((((....))))))).))).....(....)......))))....))).. ( -29.50, z-score = -1.67, R) >droYak2.chrX 18251789 86 - 21770863 --------------------------UGGCUAUGGCUAUGUCAUGUGGCUG-------GGCCAAGCGACUAAUUGAGCAAUUAGUGGCUAAAAAGGCAACAAUUGAGGCCAUUUGCUGG --------------------------.(((...((((((.....)))))).-------((((.(((.(((((((....))))))).))).....(....)......))))....))).. ( -29.30, z-score = -1.67, R) >droSec1.super_15 415394 91 - 1954846 ---------------------GAUCCGGGCUAUGGCUAUGUCAUGUGGCUG-------GGCCAAGCGACUAAUUGAGCAAUUAGUGGCUAAAAAGGCAACAAUUGAGGCCAUUUGCUGG ---------------------...((((.....((((((.....)))))).-------((((.(((.(((((((....))))))).))).....(....)......)))).....)))) ( -30.60, z-score = -1.46, R) >droSim1.chrX 7700630 91 - 17042790 ---------------------GAUCCAGGCUAUGGCUAUGUCAUGUGGCUG-------GGCCAAGCGACUAAUUGAGCAAUUAGUGGCUAAAAAGGCAACAAUUGAGGCCAUUUGCUGG ---------------------...((((.....((((((.....)))))).-------((((.(((.(((((((....))))))).))).....(....)......)))).....)))) ( -31.30, z-score = -1.69, R) >consensus _____________________G_U_CGGGCUAUGGCUAUGUCAUGUGGCUG_______GGCCAAGCGACUAAUUGAGCAAUUAGUGGCUAAAAAGGCAACAAUUGAGGCCAUUUGCUGG ...........................(((...((((((.....))))))........((((.(((.(((((((....))))))).))).....(....)......))))....))).. (-21.55 = -21.59 + 0.04)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:29:21 2011