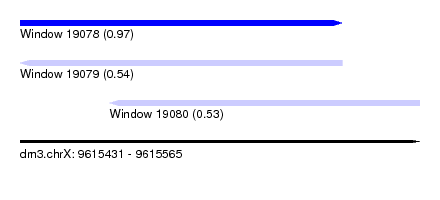
| Sequence ID | dm3.chrX |
|---|---|
| Location | 9,615,431 – 9,615,565 |
| Length | 134 |
| Max. P | 0.971032 |

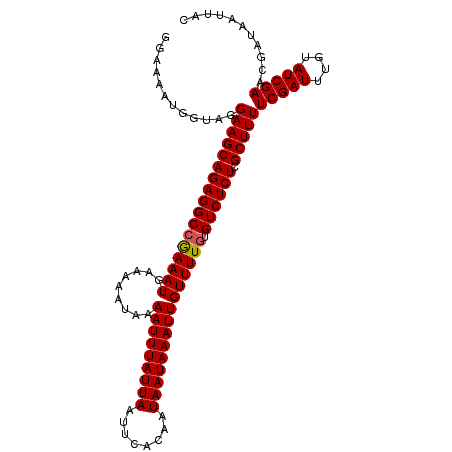
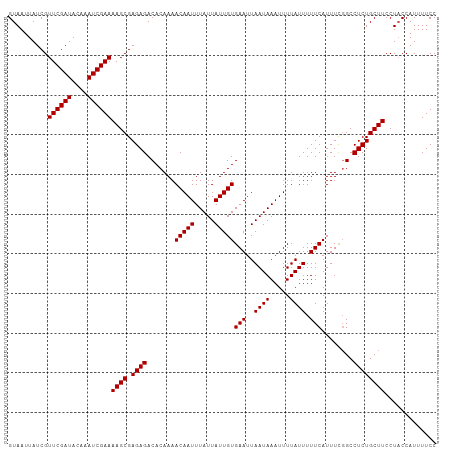
| Location | 9,615,431 – 9,615,539 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 98.70 |
| Shannon entropy | 0.02236 |
| G+C content | 0.30000 |
| Mean single sequence MFE | -23.74 |
| Consensus MFE | -22.38 |
| Energy contribution | -22.34 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.94 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.971032 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
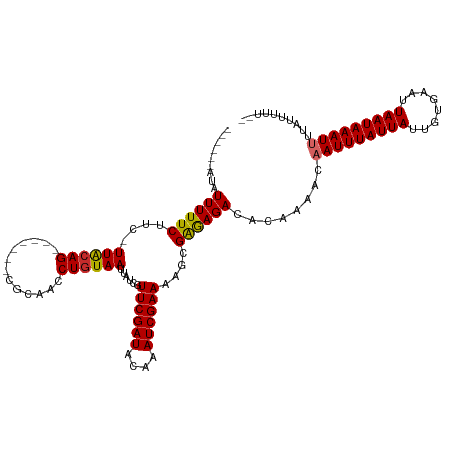
>dm3.chrX 9615431 108 + 22422827 GGAAAAUGGUAGGAAGCAGAGGCCGAAAUGAAAAAUAAAAUUUAUUAAUUCACAAUAAUAAAUUGUUUUGUGUCUCUCGCUUUUCGAUUUGUAUCGAACGAUAAUUAC .....((.((..(((((((((((((((((.........(((((((((........))))))))))))))).)))))).)))))(((((....))))))).))...... ( -24.00, z-score = -2.36, R) >droEre2.scaffold_4690 13247454 108 - 18748788 GGAAAAUGGUAGGAAGCAGAGGCAAAAAUGAAAAAUAAAAUUUAUUAAUUCACAAUAAUAAAUUGUUUUGUGUCUCUCGCUUUUCGAUUUGUAUCGAACGAUAAUUAC .....((.((..((((((((((((.......((((((..((((((((........)))))))))))))).))))))).)))))(((((....))))))).))...... ( -22.40, z-score = -2.40, R) >droYak2.chrX 18212775 108 + 21770863 GGAAAAUGGAAGGAAGCAGAGGCCAAAAUGAAAAAUAAAAUUUAUUAAUUCACAAUAAUAAAUUGUUUUGUGUCUCUCGCUUUUCGAUUUGUAUCGAACGAUAAUUAC ...((((.((..(((((((((((((((((.........(((((((((........))))))))))))))).)))))).))))))).)))).((((....))))..... ( -24.30, z-score = -2.82, R) >droSec1.super_15 378927 108 + 1954846 GGAAAAUGGUAGGAAGCAGAGGCCGAAAUGAAAAAUAAAAUUUAUUAAUUCACAAUAAUAAAUUGUUUUGUGUCUCUCGCUUUUCGAUUUGUAUCGAACGAUAAUUAC .....((.((..(((((((((((((((((.........(((((((((........))))))))))))))).)))))).)))))(((((....))))))).))...... ( -24.00, z-score = -2.36, R) >droSim1.chrX 7663335 108 + 17042790 GGAAAAUGGUAGGAAGCAGAGGCCGAAAUGAAAAAUAAAAUUUAUUAAUUCACAAUAAUAAAUUGUUUUGUGUCUCUCGCUUUUCGAUUUGUAUCGAACGAUAAUUAC .....((.((..(((((((((((((((((.........(((((((((........))))))))))))))).)))))).)))))(((((....))))))).))...... ( -24.00, z-score = -2.36, R) >consensus GGAAAAUGGUAGGAAGCAGAGGCCGAAAUGAAAAAUAAAAUUUAUUAAUUCACAAUAAUAAAUUGUUUUGUGUCUCUCGCUUUUCGAUUUGUAUCGAACGAUAAUUAC ............(((((((((((((((((.........(((((((((........))))))))))))))).)))))).)))))(((((....)))))........... (-22.38 = -22.34 + -0.04)

| Location | 9,615,431 – 9,615,539 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 98.70 |
| Shannon entropy | 0.02236 |
| G+C content | 0.30000 |
| Mean single sequence MFE | -15.60 |
| Consensus MFE | -14.90 |
| Energy contribution | -14.90 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.96 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.536703 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 9615431 108 - 22422827 GUAAUUAUCGUUCGAUACAAAUCGAAAAGCGAGAGACACAAAACAAUUUAUUAUUGUGAAUUAAUAAAUUUUAUUUUUCAUUUCGGCCUCUGCUUCCUACCAUUUUCC (((.......((((((....))))))((((.((((.(.....(((((.....)))))(((..((((.....)))).))).....)..))))))))..)))........ ( -15.00, z-score = -0.85, R) >droEre2.scaffold_4690 13247454 108 + 18748788 GUAAUUAUCGUUCGAUACAAAUCGAAAAGCGAGAGACACAAAACAAUUUAUUAUUGUGAAUUAAUAAAUUUUAUUUUUCAUUUUUGCCUCUGCUUCCUACCAUUUUCC (((.......((((((....))))))((((.((((.((.((((((((.....)))))(((..((((.....)))).))).))).)).))))))))..)))........ ( -16.00, z-score = -1.69, R) >droYak2.chrX 18212775 108 - 21770863 GUAAUUAUCGUUCGAUACAAAUCGAAAAGCGAGAGACACAAAACAAUUUAUUAUUGUGAAUUAAUAAAUUUUAUUUUUCAUUUUGGCCUCUGCUUCCUUCCAUUUUCC ..........((((((....))))))((((.((((.(.(((((((((.....)))))(((..((((.....)))).)))..))))).))))))))............. ( -17.00, z-score = -1.47, R) >droSec1.super_15 378927 108 - 1954846 GUAAUUAUCGUUCGAUACAAAUCGAAAAGCGAGAGACACAAAACAAUUUAUUAUUGUGAAUUAAUAAAUUUUAUUUUUCAUUUCGGCCUCUGCUUCCUACCAUUUUCC (((.......((((((....))))))((((.((((.(.....(((((.....)))))(((..((((.....)))).))).....)..))))))))..)))........ ( -15.00, z-score = -0.85, R) >droSim1.chrX 7663335 108 - 17042790 GUAAUUAUCGUUCGAUACAAAUCGAAAAGCGAGAGACACAAAACAAUUUAUUAUUGUGAAUUAAUAAAUUUUAUUUUUCAUUUCGGCCUCUGCUUCCUACCAUUUUCC (((.......((((((....))))))((((.((((.(.....(((((.....)))))(((..((((.....)))).))).....)..))))))))..)))........ ( -15.00, z-score = -0.85, R) >consensus GUAAUUAUCGUUCGAUACAAAUCGAAAAGCGAGAGACACAAAACAAUUUAUUAUUGUGAAUUAAUAAAUUUUAUUUUUCAUUUCGGCCUCUGCUUCCUACCAUUUUCC ..........((((((....))))))((((.((((.......(((((.....)))))(((..((((.....)))).)))........))))))))............. (-14.90 = -14.90 + 0.00)

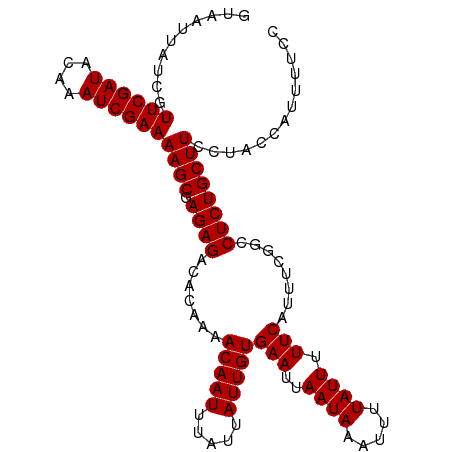
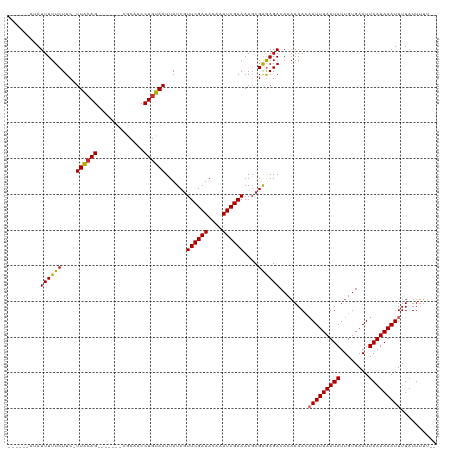
| Location | 9,615,461 – 9,615,565 |
|---|---|
| Length | 104 |
| Sequences | 9 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.35 |
| Shannon entropy | 0.18900 |
| G+C content | 0.26752 |
| Mean single sequence MFE | -16.96 |
| Consensus MFE | -13.04 |
| Energy contribution | -13.01 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.532887 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chrX 9615461 104 - 22422827 ------AUAUUUUUCUUC-UUACAG-------CGCAACCUGUAAUUAUCGUUCGAUACAAAUCGAAAAGCGAGAGACACAAAACAAUUUAUUAUUGUGAAUUAAUAAAUUUUAUUUUU-- ------.......((((.-((((((-------......))))))...(((((((((....)))))...))))))))........(((((((((........)))))))))........-- ( -16.10, z-score = -1.42, R) >droMoj3.scaffold_6473 16169886 109 - 16943266 ACUGAGGCAUUUUUCUUC-UUACAG-------CGCAACCUGUAAUUAUCAUUCGAUACAAAUCGAAAA-CGAAAGACACUAAA--AUUUAUUAUUGUGAAUUAAUAAAUUUUUUUGUGGU .........((((((...-((((((-------......))))))......((((((....))))))..-.))))))(((.(((--((((((((........)))))))))))...))).. ( -21.90, z-score = -2.45, R) >droWil1.scaffold_181096 3358564 108 - 12416693 ACCGAAACAUUUUUCUUC-UUACAGC------CACAACCUGUAAUUAUCGUUCGAUACAAAUCGAAAA-CGAAAGACACAAAA--AUUUAUUAUUGCGAAUUAAUAAAUUUUAUUUCU-- ...((((......((((.-((((((.------......))))))...(((((((((....)))))..)-)))))))....(((--((((((((........))))))))))).)))).-- ( -19.10, z-score = -3.77, R) >droPer1.super_15 126854 108 + 2181545 ---GAUAAAUUUUUCUUCUUUGGAG-------CGCAACCUGUAAUUAUCGUUCGAUACAAAUCGAAAAGCGGGAGACACAAAACAAUUUAUUAUUGUGAAUUAAUAAAUUUUAUUUUU-- ---(((((..........((((...-------(....(((((........((((((....))))))..))))).)...))))..(((((((((........))))))))))))))...-- ( -13.80, z-score = 0.49, R) >droAna3.scaffold_13248 1439393 111 - 4840945 ------ACAUUUUUCUUC-UUACAGUCCCGCAUACAACCUGUAAUUAUCGUUCGAUACAAAUCGAAAAGCGAGAGACACAAAACAAUUUAUUAUUGUGAAUUAAUAAAUUUUAUUUUU-- ------............-.....(((((((.(((.....))).......((((((....))))))..))).).))).......(((((((((........)))))))))........-- ( -17.30, z-score = -2.11, R) >droEre2.scaffold_4690 13247484 104 + 18748788 ------AUAUUUUUCUUC-UUACAG-------CGCAACCUGUAAUUAUCGUUCGAUACAAAUCGAAAAGCGAGAGACACAAAACAAUUUAUUAUUGUGAAUUAAUAAAUUUUAUUUUU-- ------.......((((.-((((((-------......))))))...(((((((((....)))))...))))))))........(((((((((........)))))))))........-- ( -16.10, z-score = -1.42, R) >droYak2.chrX 18212805 104 - 21770863 ------AUAUUUUUCUUC-UUACAG-------CACAACCUGUAAUUAUCGUUCGAUACAAAUCGAAAAGCGAGAGACACAAAACAAUUUAUUAUUGUGAAUUAAUAAAUUUUAUUUUU-- ------.......((((.-((((((-------......))))))...(((((((((....)))))...))))))))........(((((((((........)))))))))........-- ( -16.10, z-score = -1.96, R) >droSec1.super_15 378957 104 - 1954846 ------AUAUUUUUCUUC-UUACAG-------CGCAACCUGUAAUUAUCGUUCGAUACAAAUCGAAAAGCGAGAGACACAAAACAAUUUAUUAUUGUGAAUUAAUAAAUUUUAUUUUU-- ------.......((((.-((((((-------......))))))...(((((((((....)))))...))))))))........(((((((((........)))))))))........-- ( -16.10, z-score = -1.42, R) >droSim1.chrX 7663365 104 - 17042790 ------AUAUUUUUCUUC-UUACAG-------CGCAACCUGUAAUUAUCGUUCGAUACAAAUCGAAAAGCGAGAGACACAAAACAAUUUAUUAUUGUGAAUUAAUAAAUUUUAUUUUU-- ------.......((((.-((((((-------......))))))...(((((((((....)))))...))))))))........(((((((((........)))))))))........-- ( -16.10, z-score = -1.42, R) >consensus ______AUAUUUUUCUUC_UUACAG_______CGCAACCUGUAAUUAUCGUUCGAUACAAAUCGAAAAGCGAGAGACACAAAACAAUUUAUUAUUGUGAAUUAAUAAAUUUUAUUUUU__ .........((((((....((((((.............))))))......((((((....))))))....))))))........(((((((((........))))))))).......... (-13.04 = -13.01 + -0.03)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:29:18 2011