| Sequence ID | dm3.chrX |
|---|---|
| Location | 9,606,816 – 9,606,931 |
| Length | 115 |
| Max. P | 0.840095 |

| Location | 9,606,816 – 9,606,931 |
|---|---|
| Length | 115 |
| Sequences | 7 |
| Columns | 121 |
| Reading direction | reverse |
| Mean pairwise identity | 68.93 |
| Shannon entropy | 0.58166 |
| G+C content | 0.42213 |
| Mean single sequence MFE | -24.97 |
| Consensus MFE | -11.50 |
| Energy contribution | -11.04 |
| Covariance contribution | -0.46 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.840095 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
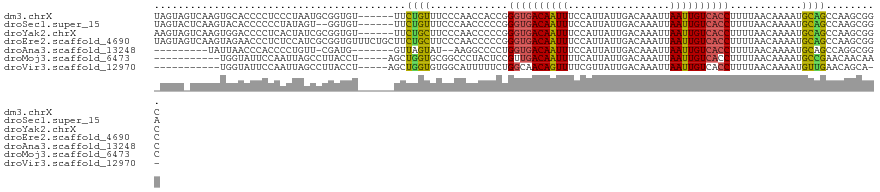
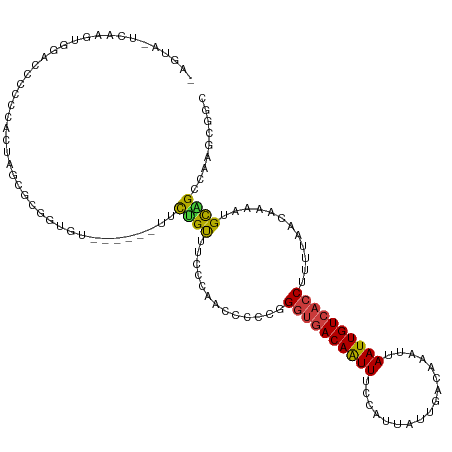
>dm3.chrX 9606816 115 - 22422827 UAGUAGUCAAGUGCACCCCUCCCUAAUGCGGUGU------UUCUGUUUCCCAACCACCGGGUGACAAUUUCCAUUAUUGACAAAUUAAUUGUCACCUUUUAACAAAAUGCAGCCAAGCGGC ...........((((.............(((((.------..............)))))((((((((((.................))))))))))...........))))(((....))) ( -24.29, z-score = -0.70, R) >droSec1.super_15 370558 113 - 1954846 UAGUACUCAAGUACACCCCCCUAUAGU--GGUGU------UUCUGUUUCCCAACCCCCGGGUGACAAUUUCCAUUAUUGACAAAUUAAUUGUCACCUUUUAACAAAAUGCAGCCAAGCGGA ..((((....)))).....((.(((((--((...------...((((.(((.......))).))))....)))))))((((((.....))))))..............((......)))). ( -23.50, z-score = -1.07, R) >droYak2.chrX 18202723 115 - 21770863 AAGUAGUCAAGUGGACCCCUCACUAUCGCGGUGU------UUCUGCUUCCCAACCCCCGGGUGACAAUUUCCAUUAUUGACAAAUUAAUUGUCACCUUUUAACAAAAUGCAGCCAAGCGGC .........(((((.....))))).((((..((.------..((((............(((((((((((.................)))))))))))...........)))).)).)))). ( -25.83, z-score = -0.79, R) >droEre2.scaffold_4690 13238828 121 + 18748788 UAGUAGUCAAGUAGAACCCUCUCCAUCGCGGUGUUUCUGCUUCUGCUUCCCAACCCCCGGGUGACAAUUUCCAUUAUUGACAAAUUAAUUGUCACCUUUUAACAAAAUGCAGCCAAGCGGC .(((((..(((((((((((.(......).)).).)))))))))))))...........(((((((((((.................)))))))))))..............(((....))) ( -29.03, z-score = -2.15, R) >droAna3.scaffold_13248 1429930 102 - 4840945 ---------UAUUAACCCACCCCUGUU-CGAUG-------GUUAGUAU--AAGGCCCCUGGUGACAAUUUCCAUUAUUGACAAAUUAAUUGUCACCUUUUAACAAAAUGCAGCCAGGCGGC ---------((((((((..........-....)-------))))))).--...(((((((((..((.(((.......((((((.....)))))).........))).))..)))))).))) ( -24.93, z-score = -1.46, R) >droMoj3.scaffold_6473 16154960 105 - 16943266 -----------UGGUAUUCCAAUUAGCCUUACCU-----AGCUGGUGCGGCCCUACUCCGUUGACAAUUUUCAUUAUUGACAAAUUAAUUGUCACCUUUUAACAAAAUGCCGAACAACAAC -----------.((((((.......(((.((((.-----....)))).)))........(.((((((((.(((....)))......)))))))).).........)))))).......... ( -22.00, z-score = -2.33, R) >droVir3.scaffold_12970 2677846 103 + 11907090 -----------UGGUAUUCCAAUUAGCCUUACCU-----AGCUGGUGUGGCAUUUUUCUGGCAACAGUUUUCGUUAUUGACAAAUUAAUUGUCACCUUUUAACAAAAUGUUGAACAGCA-- -----------.((((.............)))).-----.((((...(((((((((.(((....))).....((((.((((((.....)))))).....)))))))))))))..)))).-- ( -25.22, z-score = -1.78, R) >consensus _AGUA_UCAAGUGGACCCCCCACUAGCGCGGUGU______UUCUGUUUCCCAACCCCCGGGUGACAAUUUCCAUUAUUGACAAAUUAAUUGUCACCUUUUAACAAAAUGCAGCCAAGCGGC ..........................................((((.............((((((((((.................))))))))))............))))......... (-11.50 = -11.04 + -0.46)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:29:15 2011