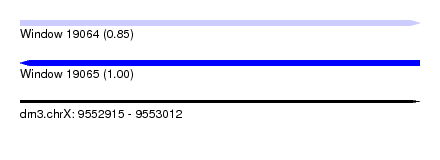
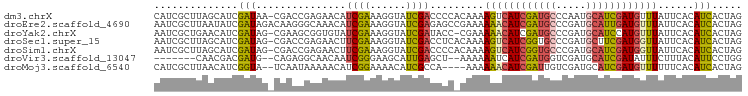
| Sequence ID | dm3.chrX |
|---|---|
| Location | 9,552,915 – 9,553,012 |
| Length | 97 |
| Max. P | 0.997228 |

| Location | 9,552,915 – 9,553,012 |
|---|---|
| Length | 97 |
| Sequences | 7 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 73.90 |
| Shannon entropy | 0.51975 |
| G+C content | 0.43926 |
| Mean single sequence MFE | -28.84 |
| Consensus MFE | -11.18 |
| Energy contribution | -12.00 |
| Covariance contribution | 0.82 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.848899 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
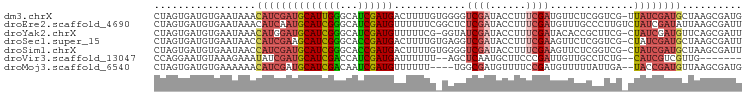
>dm3.chrX 9552915 97 + 22422827 CUAGUGAUGUGAAUAAACAUCGAUGCAUUGGGCAUCGAUGACUUUUGUGGGGUCGAUACCUUUCGAUGUUCUCGGUCG-UUAUCGAUGCUAAGCGAUG ....((((((......)))))).(((....((((((((((((..(((.((((((((......))))).))).)))..)-)))))))))))..)))... ( -34.20, z-score = -3.15, R) >droEre2.scaffold_4690 13183332 98 - 18748788 CUAGUGAUGUGAAUAAACAUCAAUGCAUCGGGCAUCGAUGUUUUUUCGGCUCUCGAUACCUUUCGAUGUUUGCCCUUGUCUAUCGAUAUUAAGCGAUU ....(((((((((.(((((((.((((.....)))).))))))).)))(((.(((((......)))).)...)))...........))))))....... ( -24.40, z-score = -1.30, R) >droYak2.chrX 18143520 96 + 21770863 CUAGUGAUGUGAAUAAACAUGGAUGCAUCGGGCAUCGAUGUUUUUCG-GGUAUCGAUACCUUUCGAUACACCGCUUCG-CUAUCGAUGUUCAGCGAUU .((((((.(((((.((((((.(((((.....))))).)))))))))(-((((((((......))))))).)))).)))-)))(((........))).. ( -35.50, z-score = -3.69, R) >droSec1.super_15 316492 97 + 1954846 CUAGUGAUGUGAAUAACCAUCGAAGCAUCGGGCACCGAUGACUUUUGUGAGGUCGAUACCUUUCGAAGUUCUCGGUCG-CUAUCGAUGCUAAGCGAUU ...((((((........))))..(((((((((((((((.((((((...(((((....)))))..)))))).))))).)-))..)))))))..)).... ( -34.20, z-score = -2.87, R) >droSim1.chrX 7606080 97 + 17042790 CUAGUGAUGUGAAUAACCAUCGAUGCAUCGGGCACCGAUGACUUUUGUGGGGUCGAUACCUUUCGAAGUUCUCGGUCG-CUAUCGAUGCUAAGCGAUU ..................((((..((((((((((((((.((((((.(..((((....))))..))))))).))))).)-))..))))))....)))). ( -32.40, z-score = -2.00, R) >droVir3.scaffold_13047 17643088 87 - 19223366 CCAGGAAUGUAAAGAAAUAUCGAUGCAUCGACCAUCGAUGAUUUUUU--AGCUCAAUGCUUCCCGAUUGUUGCCUCUG--CAUCGUCGUUG------- ...((((..(((((((.((((((((.......)))))))).))))))--)((.....))))))((((...(((....)--))..))))...------- ( -21.20, z-score = -1.52, R) >droMoj3.scaffold_6540 8179763 92 - 34148556 CUAGUGAUGUGAAAAAACAUCGAUGCAUCGACAAUCGAUGUUUUUU----UGGCGAUGUUUUCCGAUGUUUUUAUUGA--UACCGAUGUUAAGCGAUG .......(((..((((((((((..((((((.(((...........)----)).))))))....)))))))))).((((--((....)))))))))... ( -20.00, z-score = -0.46, R) >consensus CUAGUGAUGUGAAUAAACAUCGAUGCAUCGGGCAUCGAUGAUUUUUG_GGGGUCGAUACCUUUCGAUGUUCUCCGUCG_CUAUCGAUGUUAAGCGAUU .................((((((((((((((...))))))............((((......))))..............)))))))).......... (-11.18 = -12.00 + 0.82)
| Location | 9,552,915 – 9,553,012 |
|---|---|
| Length | 97 |
| Sequences | 7 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 73.90 |
| Shannon entropy | 0.51975 |
| G+C content | 0.43926 |
| Mean single sequence MFE | -25.15 |
| Consensus MFE | -13.67 |
| Energy contribution | -13.57 |
| Covariance contribution | -0.09 |
| Combinations/Pair | 1.42 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.06 |
| SVM RNA-class probability | 0.997228 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
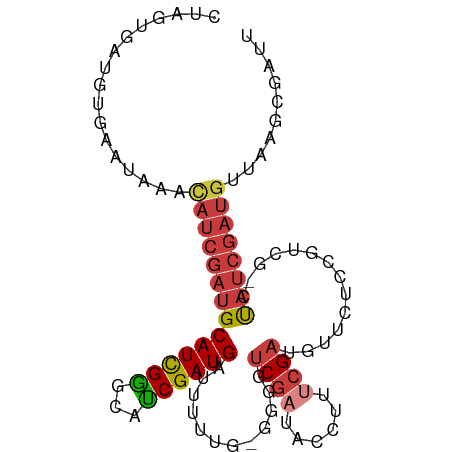
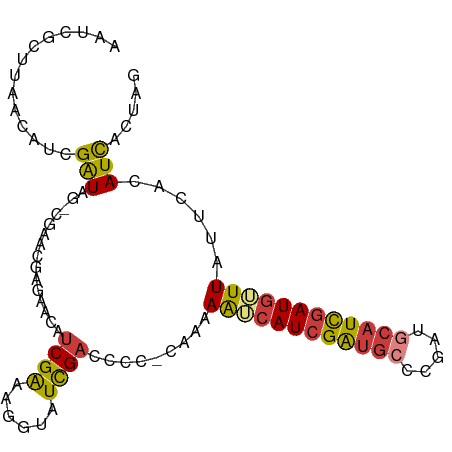
>dm3.chrX 9552915 97 - 22422827 CAUCGCUUAGCAUCGAUAA-CGACCGAGAACAUCGAAAGGUAUCGACCCCACAAAAGUCAUCGAUGCCCAAUGCAUCGAUGUUUAUUCACAUCACUAG ....((...((((((((.(-(...(((.....)))...(((....)))........)).)))))))).....))...(((((......)))))..... ( -23.20, z-score = -2.88, R) >droEre2.scaffold_4690 13183332 98 + 18748788 AAUCGCUUAAUAUCGAUAGACAAGGGCAAACAUCGAAAGGUAUCGAGAGCCGAAAAAACAUCGAUGCCCGAUGCAUUGAUGUUUAUUCACAUCACUAG ....((((....(((((......(......)(((....)))))))))))).(((.((((((((((((.....)))))))))))).))).......... ( -27.00, z-score = -3.28, R) >droYak2.chrX 18143520 96 - 21770863 AAUCGCUGAACAUCGAUAG-CGAAGCGGUGUAUCGAAAGGUAUCGAUACC-CGAAAAACAUCGAUGCCCGAUGCAUCCAUGUUUAUUCACAUCACUAG ..((((((........)))-)))...((.(((((((......))))))))-)(((((((((.(((((.....))))).)))))).))).......... ( -30.30, z-score = -3.61, R) >droSec1.super_15 316492 97 - 1954846 AAUCGCUUAGCAUCGAUAG-CGACCGAGAACUUCGAAAGGUAUCGACCUCACAAAAGUCAUCGGUGCCCGAUGCUUCGAUGGUUAUUCACAUCACUAG ........(((((((...(-(.(((((..((((....((((....)))).....))))..))))))).)))))))..((((........))))..... ( -29.40, z-score = -2.78, R) >droSim1.chrX 7606080 97 - 17042790 AAUCGCUUAGCAUCGAUAG-CGACCGAGAACUUCGAAAGGUAUCGACCCCACAAAAGUCAUCGGUGCCCGAUGCAUCGAUGGUUAUUCACAUCACUAG .........((((((...(-(.(((((..((((.....(((....)))......))))..))))))).))))))...((((........))))..... ( -27.80, z-score = -2.49, R) >droVir3.scaffold_13047 17643088 87 + 19223366 -------CAACGACGAUG--CAGAGGCAACAAUCGGGAAGCAUUGAGCU--AAAAAAUCAUCGAUGGUCGAUGCAUCGAUAUUUCUUUACAUUCCUGG -------......(((((--((..(((....((((((((((.....)))--......)).))))).)))..))))))).................... ( -21.60, z-score = -1.54, R) >droMoj3.scaffold_6540 8179763 92 + 34148556 CAUCGCUUAACAUCGGUA--UCAAUAAAAACAUCGGAAAACAUCGCCA----AAAAAACAUCGAUUGUCGAUGCAUCGAUGUUUUUUCACAUCACUAG ..............(((.--.....(((((((((((....(((((.((----(...........))).)))))..))))))))))).......))).. ( -16.72, z-score = -2.05, R) >consensus AAUCGCUUAACAUCGAUAG_CGAACGAGAACAUCGAAAGGUAUCGACCCC_CAAAAAUCAUCGAUGCCCGAUGCAUCGAUGUUUAUUCACAUCACUAG ..............(((...............((((......)))).........((((((((((((.....))))))))))))......)))..... (-13.67 = -13.57 + -0.09)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:29:06 2011