| Sequence ID | dm3.chrX |
|---|---|
| Location | 9,540,875 – 9,540,967 |
| Length | 92 |
| Max. P | 0.995415 |

| Location | 9,540,875 – 9,540,967 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 76.39 |
| Shannon entropy | 0.45566 |
| G+C content | 0.42550 |
| Mean single sequence MFE | -24.82 |
| Consensus MFE | -17.44 |
| Energy contribution | -17.34 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.43 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.80 |
| SVM RNA-class probability | 0.995415 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
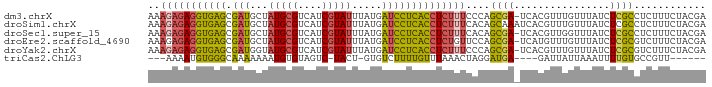
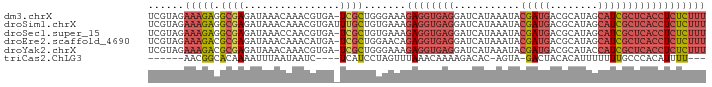
>dm3.chrX 9540875 92 + 22422827 AAAGAGAGGUGAGCGAUGCUAUGCGUCAUCGUAUUUAUGAUCCUCACCUCUUUCCCAGCGA-UCACGUUUGUUUAUCUCGCCUCUUUCUACGA ..(((((((((((....((...((((.(((((......((........)).......))))-).))))..))....)))))))))))...... ( -26.32, z-score = -2.72, R) >droSim1.chrX 7593013 93 + 17042790 AAAGAGAGGUGAGCGAUGCUAUGCGUCAUCGUAUUUAUGAUCCUCACCUCUUUCACAGCAAAUCACGUUUGUUUAUCUCGCCUCUUUCUACGA ..(((((((((((.(((((...)))))(((((....))))).)))))))))))...(((((((...))))))).................... ( -24.70, z-score = -2.46, R) >droSec1.super_15 304552 92 + 1954846 AAAGAGAGGUGAGCGAUGCUAUGCGUCAUCGUAUUUAUGAUCCUCACCUCUUUCACAGCGA-UCACGUUGGUUUAUCUCGCCUCUUUCUACGA ..(((((((((((....((((.((((.(((((......((........)).......))))-).))))))))....)))))))))))...... ( -27.82, z-score = -2.47, R) >droEre2.scaffold_4690 13171431 92 - 18748788 AAAGAGAGGUGAGCGAUGCUAUGCGUCAUCGUAUUUAUGAUCCUCACCUCUGUUCCAGCGA-UCAUGUUUGUUUAUCUCGCGUCUUUCUACGA .((.(((((((((.(((((...)))))(((((....))))).))))))))).))...((((-..(((......))).))))............ ( -26.50, z-score = -2.00, R) >droYak2.chrX 18131413 92 + 21770863 AAAGAGAGGUGAGCGAUGGUAUGCGUCAUCGUAUUUAUGAUCCUCACCUCUUUCCCAGCGA-UCACGUUUGUUUAUCUCGCGUCUUUCUACGA ..(((((((((((.(((.(((((((....))))..))).))))))))))))))....((((-...............))))............ ( -28.06, z-score = -2.53, R) >triCas2.ChLG3 19966696 78 - 32080666 ---AAAAUGUGGGCAAAAAAAUGUGUAGUC-UACU-GUGUCUUUUGUUUAAACUAGGAUGA----GAUUAUUAAAUUUUGUGCCGUU------ ---........((((..(((((..((((((-(...-..(((((..((....)).))))).)----))))))...))))).))))...------ ( -15.50, z-score = -2.05, R) >consensus AAAGAGAGGUGAGCGAUGCUAUGCGUCAUCGUAUUUAUGAUCCUCACCUCUUUCACAGCGA_UCACGUUUGUUUAUCUCGCCUCUUUCUACGA ..(((((((((((.(((...(((((....))))).....))))))))))))))....((((................))))............ (-17.44 = -17.34 + -0.10)
| Location | 9,540,875 – 9,540,967 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 76.39 |
| Shannon entropy | 0.45566 |
| G+C content | 0.42550 |
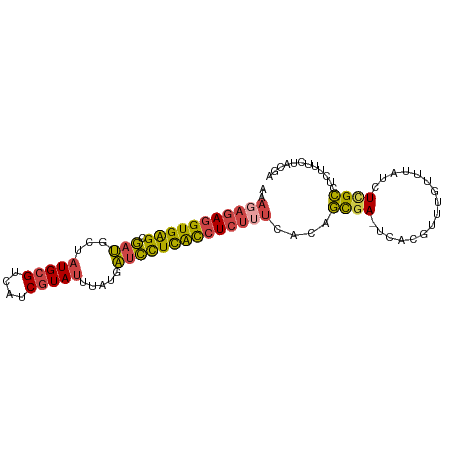
| Mean single sequence MFE | -21.28 |
| Consensus MFE | -11.89 |
| Energy contribution | -14.08 |
| Covariance contribution | 2.20 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.676607 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
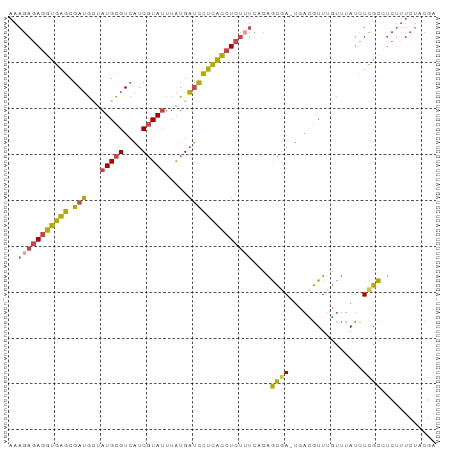
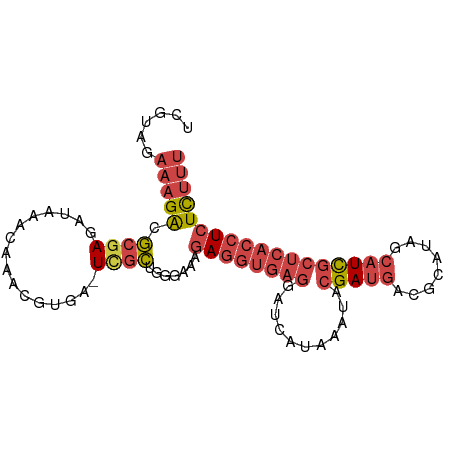
>dm3.chrX 9540875 92 - 22422827 UCGUAGAAAGAGGCGAGAUAAACAAACGUGA-UCGCUGGGAAAGAGGUGAGGAUCAUAAAUACGAUGACGCAUAGCAUCGCUCACCUCUCUUU ......((((((((((.....((....))..-)))))......((((((((...........(((((.(.....))))))))))))))))))) ( -25.20, z-score = -1.68, R) >droSim1.chrX 7593013 93 - 17042790 UCGUAGAAAGAGGCGAGAUAAACAAACGUGAUUUGCUGUGAAAGAGGUGAGGAUCAUAAAUACGAUGACGCAUAGCAUCGCUCACCUCUCUUU ......(((((((((((....((....))..))))))......((((((((...........(((((.(.....))))))))))))))))))) ( -23.30, z-score = -1.20, R) >droSec1.super_15 304552 92 - 1954846 UCGUAGAAAGAGGCGAGAUAAACCAACGUGA-UCGCUGUGAAAGAGGUGAGGAUCAUAAAUACGAUGACGCAUAGCAUCGCUCACCUCUCUUU ......((((((((((.....((....))..-)))))......((((((((...........(((((.(.....))))))))))))))))))) ( -24.60, z-score = -1.30, R) >droEre2.scaffold_4690 13171431 92 + 18748788 UCGUAGAAAGACGCGAGAUAAACAAACAUGA-UCGCUGGAACAGAGGUGAGGAUCAUAAAUACGAUGACGCAUAGCAUCGCUCACCUCUCUUU ......(((((.((((...............-)))).......((((((((...........(((((.(.....))))))))))))))))))) ( -22.76, z-score = -1.48, R) >droYak2.chrX 18131413 92 - 21770863 UCGUAGAAAGACGCGAGAUAAACAAACGUGA-UCGCUGGGAAAGAGGUGAGGAUCAUAAAUACGAUGACGCAUACCAUCGCUCACCUCUCUUU ((.(((...((((((...........)))).-)).))).)).(((((((((...........(((((........)))))))))))))).... ( -25.40, z-score = -2.00, R) >triCas2.ChLG3 19966696 78 + 32080666 ------AACGGCACAAAAUUUAAUAAUC----UCAUCCUAGUUUAAACAAAAGACAC-AGUA-GACUACACAUUUUUUUGCCCACAUUUU--- ------...((((.((((..........----........((((.......))))..-.(((-...)))....)))).))))........--- ( -6.40, z-score = -0.95, R) >consensus UCGUAGAAAGACGCGAGAUAAACAAACGUGA_UCGCUGGGAAAGAGGUGAGGAUCAUAAAUACGAUGACGCAUAGCAUCGCUCACCUCUCUUU ......(((((.((((.....((....))...)))).......((((((((...........(((((........)))))))))))))))))) (-11.89 = -14.08 + 2.20)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:29:02 2011