
| Sequence ID | dm3.chrX |
|---|---|
| Location | 9,538,637 – 9,538,908 |
| Length | 271 |
| Max. P | 0.892516 |

| Location | 9,538,637 – 9,538,755 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.75 |
| Shannon entropy | 0.12655 |
| G+C content | 0.42005 |
| Mean single sequence MFE | -35.12 |
| Consensus MFE | -25.60 |
| Energy contribution | -25.84 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.892516 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 9538637 118 + 22422827 CCAAGUGAAUUUACGACAAUUGGUUAAAUUACGGAACUAGUUCUGUCCCUCUGACGCACAUAAAGUGGUUAACUGGUUUCCGCUUUGCCAUUGCCC--AUUUGAUUCAUGGGCAUUGGCA ..((((((((((((........).))))))..((((((((((..(((.....))).(((.....)))...)))))))))))))))(((((.(((((--((.......))))))).))))) ( -36.30, z-score = -2.56, R) >droSim1.chrX 7590769 118 + 17042790 CCAAGUGAAUUUACGACAAUUGGUUAAAUUACGGAACUAGUUCUGUCACUCUGACACACAUAAAGUGGUCAACUGGUUUCCGCUUUGCCAUUGCCC--AUUUGAUUCAUGGGCAUUGGCA ..((((((((((((........).))))))..((((((((((.((((.....))))(((.....)))...)))))))))))))))(((((.(((((--((.......))))))).))))) ( -38.20, z-score = -3.31, R) >droSec1.super_15 302345 118 + 1954846 CCAAGUGAAUUUACGACAAUUGGUUAAAUUACGGAACUAGUUCUGUCACUCUGACACACAUAAAGUGGUCAACUGGUUUCCGCUUUGCCAUUGCCC--AUUUGAUUCAUGGGCAUUGGCA ..((((((((((((........).))))))..((((((((((.((((.....))))(((.....)))...)))))))))))))))(((((.(((((--((.......))))))).))))) ( -38.20, z-score = -3.31, R) >droEre2.scaffold_4690 13169181 116 - 18748788 CCAAGUGAAUUUACGACAAUUGCUUCAAUUACGGAACUAGUUCUGUCACUCUGAUGCACAUAAAGUGGUCAACUGGUUUCCGCUUUGCCAUUGCCC--AUUUGAUUCAUAGACAUUGG-- (((((((((((.........(((.(((...((((((....)))))).....))).)))....((((((.(((.((((.........))))))).))--))))))))))).....))))-- ( -26.00, z-score = -0.78, R) >droYak2.chrX 18129126 118 + 21770863 GCAAGUGAAUUUACGACAAUUGGUUAAAUUACGGAACUAGUUCGGCCACUCUGACACACAUUAAGUGGUCAACUAGUUUCCGCUUUGCCAUUGCUUUGAUUUGAUUCAUGGGCAUUGA-- ((..(((((((..(((((((.(((.......(((((((((((.(((((((.(((......)))))))))))))))).)))))....)))))))..)))....)))))))..)).....-- ( -36.90, z-score = -3.65, R) >consensus CCAAGUGAAUUUACGACAAUUGGUUAAAUUACGGAACUAGUUCUGUCACUCUGACACACAUAAAGUGGUCAACUGGUUUCCGCUUUGCCAUUGCCC__AUUUGAUUCAUGGGCAUUGGCA ..(((((..........((((.....))))..(((((((((..........((((.(((.....))))))))))))))))))))).((((.(((((.............))))).)))). (-25.60 = -25.84 + 0.24)

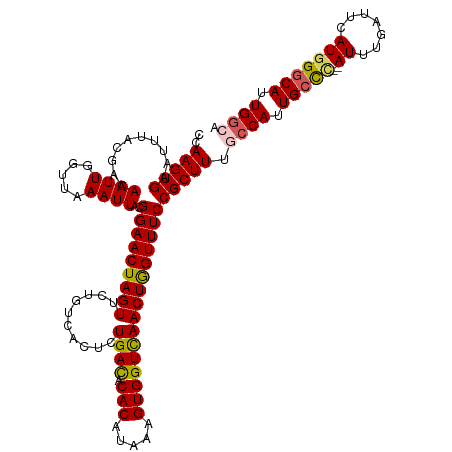

| Location | 9,538,637 – 9,538,755 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.75 |
| Shannon entropy | 0.12655 |
| G+C content | 0.42005 |
| Mean single sequence MFE | -33.84 |
| Consensus MFE | -23.28 |
| Energy contribution | -25.04 |
| Covariance contribution | 1.76 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.668663 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 9538637 118 - 22422827 UGCCAAUGCCCAUGAAUCAAAU--GGGCAAUGGCAAAGCGGAAACCAGUUAACCACUUUAUGUGCGUCAGAGGGACAGAACUAGUUCCGUAAUUUAACCAAUUGUCGUAAAUUCACUUGG (((((.(((((((.......))--))))).)))))..((((((...((((...(((.....))).(((.....)))..))))..)))))).......((((.((.........)).)))) ( -37.00, z-score = -3.10, R) >droSim1.chrX 7590769 118 - 17042790 UGCCAAUGCCCAUGAAUCAAAU--GGGCAAUGGCAAAGCGGAAACCAGUUGACCACUUUAUGUGUGUCAGAGUGACAGAACUAGUUCCGUAAUUUAACCAAUUGUCGUAAAUUCACUUGG (((((.(((((((.......))--))))).)))))..((((((.....((((((((.....))).)))))(((......)))..)))))).......((((.((.........)).)))) ( -39.40, z-score = -3.86, R) >droSec1.super_15 302345 118 - 1954846 UGCCAAUGCCCAUGAAUCAAAU--GGGCAAUGGCAAAGCGGAAACCAGUUGACCACUUUAUGUGUGUCAGAGUGACAGAACUAGUUCCGUAAUUUAACCAAUUGUCGUAAAUUCACUUGG (((((.(((((((.......))--))))).)))))..((((((.....((((((((.....))).)))))(((......)))..)))))).......((((.((.........)).)))) ( -39.40, z-score = -3.86, R) >droEre2.scaffold_4690 13169181 116 + 18748788 --CCAAUGUCUAUGAAUCAAAU--GGGCAAUGGCAAAGCGGAAACCAGUUGACCACUUUAUGUGCAUCAGAGUGACAGAACUAGUUCCGUAAUUGAAGCAAUUGUCGUAAAUUCACUUGG --(((((((((((.......))--)))))(((((...((((((...((((...((((((((....)).))))))....))))..))))))(((((...))))))))))........)))) ( -26.10, z-score = -0.22, R) >droYak2.chrX 18129126 118 - 21770863 --UCAAUGCCCAUGAAUCAAAUCAAAGCAAUGGCAAAGCGGAAACUAGUUGACCACUUAAUGUGUGUCAGAGUGGCCGAACUAGUUCCGUAAUUUAACCAAUUGUCGUAAAUUCACUUGC --....(((...(((......)))..)))(((((((.((((((.(((((((.((((((..((.....)))))))).).))))))))))))...........)))))))............ ( -27.30, z-score = -1.39, R) >consensus UGCCAAUGCCCAUGAAUCAAAU__GGGCAAUGGCAAAGCGGAAACCAGUUGACCACUUUAUGUGUGUCAGAGUGACAGAACUAGUUCCGUAAUUUAACCAAUUGUCGUAAAUUCACUUGG (((((.(((((.............))))).)))))..((((((.....((((((((.....))).)))))(((......)))..)))))).......((((.((.........)).)))) (-23.28 = -25.04 + 1.76)

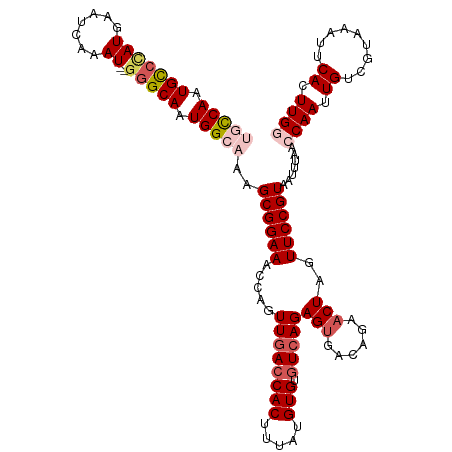

| Location | 9,538,677 – 9,538,793 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 86.77 |
| Shannon entropy | 0.22788 |
| G+C content | 0.43967 |
| Mean single sequence MFE | -35.61 |
| Consensus MFE | -20.34 |
| Energy contribution | -21.18 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.753119 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 9538677 116 + 22422827 UUCUGUCCCUCUGACGCACAUAAAGUGGUUAACUGGUUUCCGCUUUGCCAUUGCCC--AUUUGAUUCAUGGGCAUUGGCAUUGGAUAGGAUUGCCAACGGCUUGCAGAUCAAUAAAAC .(((((.((..((((.(((.....)))))))..((((.(((.((.(((((.(((((--((.......))))))).)))))..))...)))..))))..))...))))).......... ( -40.20, z-score = -2.99, R) >droSim1.chrX 7590809 116 + 17042790 UUCUGUCACUCUGACACACAUAAAGUGGUCAACUGGUUUCCGCUUUGCCAUUGCCC--AUUUGAUUCAUGGGCAUUGGCAUUAAAUAGGAUUGCCAACGGCGUGCAGAUCAAUAAAAC .((((.(((.(((((.(((.....))))))...((((.(((....(((((.(((((--((.......))))))).))))).......)))..))))..)).))))))).......... ( -42.30, z-score = -4.43, R) >droSec1.super_15 302385 116 + 1954846 UUCUGUCACUCUGACACACAUAAAGUGGUCAACUGGUUUCCGCUUUGCCAUUGCCC--AUUUGAUUCAUGGGCAUUGGCAUUAAAUAGGAUUGCCAACGGCGUGCAGAUCAAUAAAAC .((((.(((.(((((.(((.....))))))...((((.(((....(((((.(((((--((.......))))))).))))).......)))..))))..)).))))))).......... ( -42.30, z-score = -4.43, R) >droEre2.scaffold_4690 13169221 112 - 18748788 UUCUGUCACUCUGAUGCACAUAAAGUGGUCAACUGGUUUCCGCUUUGCCAUUGCCC--AUUUGAUUCAUAGACAUUGGUAAGGACUGAG----CCAACGACUUGCAGAUUAAUAAAAC .(((((.(.((((((.(((.....)))))))..((((((...((((((((.((...--..............)).))))))))...)))----)))..)).).))))).......... ( -25.53, z-score = -0.54, R) >droYak2.chrX 18129166 113 + 21770863 UUCGGCCACUCUGACACACAUUAAGUGGUCAACUAGUUUCCGCUUUGCCAUUGCUUUGAUUUGAUUCAUGGGCAUUGAAUUGGACUGC-----CCAACGGCUUGCAGAUCAAUAAAAC ...(((((((.(((......))))))))))........((.((...(((.......(((......)))((((((...........)))-----)))..)))..)).)).......... ( -27.70, z-score = -0.72, R) >consensus UUCUGUCACUCUGACACACAUAAAGUGGUCAACUGGUUUCCGCUUUGCCAUUGCCC__AUUUGAUUCAUGGGCAUUGGCAUUGAAUAGGAUUGCCAACGGCUUGCAGAUCAAUAAAAC .(((((....(((........(((((((...........)))))))((((.(((((.............))))).))))..................)))...))))).......... (-20.34 = -21.18 + 0.84)

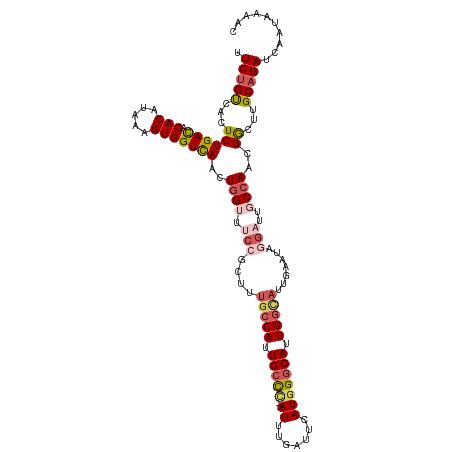
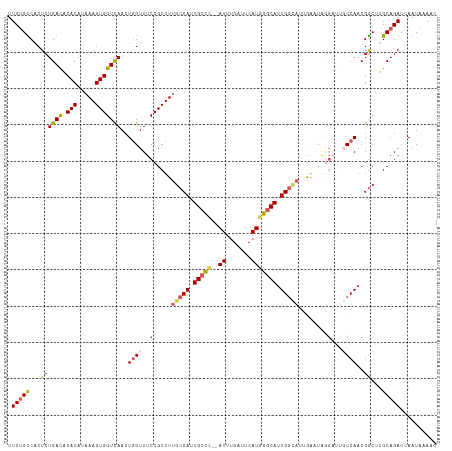
| Location | 9,538,677 – 9,538,793 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 86.77 |
| Shannon entropy | 0.22788 |
| G+C content | 0.43967 |
| Mean single sequence MFE | -36.30 |
| Consensus MFE | -24.26 |
| Energy contribution | -25.10 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.847236 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 9538677 116 - 22422827 GUUUUAUUGAUCUGCAAGCCGUUGGCAAUCCUAUCCAAUGCCAAUGCCCAUGAAUCAAAU--GGGCAAUGGCAAAGCGGAAACCAGUUAACCACUUUAUGUGCGUCAGAGGGACAGAA ((((..(((((((((..(((...)))............(((((.(((((((.......))--))))).)))))..)))))...........(((.....)))..))))..)))).... ( -37.60, z-score = -2.29, R) >droSim1.chrX 7590809 116 - 17042790 GUUUUAUUGAUCUGCACGCCGUUGGCAAUCCUAUUUAAUGCCAAUGCCCAUGAAUCAAAU--GGGCAAUGGCAAAGCGGAAACCAGUUGACCACUUUAUGUGUGUCAGAGUGACAGAA ..........(((((((.(((((((....)).......(((((.(((((((.......))--))))).))))).))))).......((((((((.....))).))))).))).)))). ( -42.40, z-score = -4.01, R) >droSec1.super_15 302385 116 - 1954846 GUUUUAUUGAUCUGCACGCCGUUGGCAAUCCUAUUUAAUGCCAAUGCCCAUGAAUCAAAU--GGGCAAUGGCAAAGCGGAAACCAGUUGACCACUUUAUGUGUGUCAGAGUGACAGAA ..........(((((((.(((((((....)).......(((((.(((((((.......))--))))).))))).))))).......((((((((.....))).))))).))).)))). ( -42.40, z-score = -4.01, R) >droEre2.scaffold_4690 13169221 112 + 18748788 GUUUUAUUAAUCUGCAAGUCGUUGG----CUCAGUCCUUACCAAUGUCUAUGAAUCAAAU--GGGCAAUGGCAAAGCGGAAACCAGUUGACCACUUUAUGUGCAUCAGAGUGACAGAA ..........((((..(.((((..(----((...(((((.(((.(((((((.......))--))))).))).))...)))....)))..))(((.....))).....)).)..)))). ( -27.00, z-score = -0.31, R) >droYak2.chrX 18129166 113 - 21770863 GUUUUAUUGAUCUGCAAGCCGUUGG-----GCAGUCCAAUUCAAUGCCCAUGAAUCAAAUCAAAGCAAUGGCAAAGCGGAAACUAGUUGACCACUUAAUGUGUGUCAGAGUGGCCGAA (((((.(((.....)))((((((((-----(((...........))))).(((......)))....)))))))))))((..(((...(((((((.....))).)))).)))..))... ( -32.10, z-score = -1.37, R) >consensus GUUUUAUUGAUCUGCAAGCCGUUGGCAAUCCUAUUCAAUGCCAAUGCCCAUGAAUCAAAU__GGGCAAUGGCAAAGCGGAAACCAGUUGACCACUUUAUGUGUGUCAGAGUGACAGAA (((.......(((((..((((((((((...........)))))))((((.............))))...)))...)))))......((((((((.....))).)))))...))).... (-24.26 = -25.10 + 0.84)

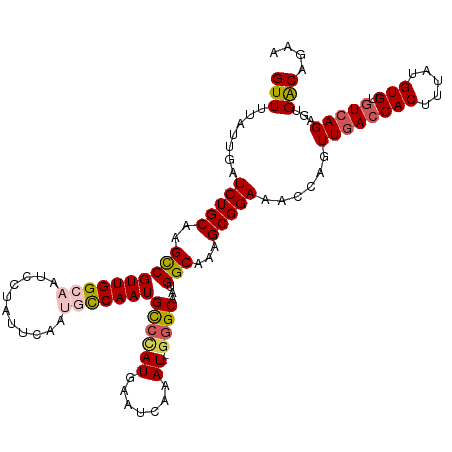
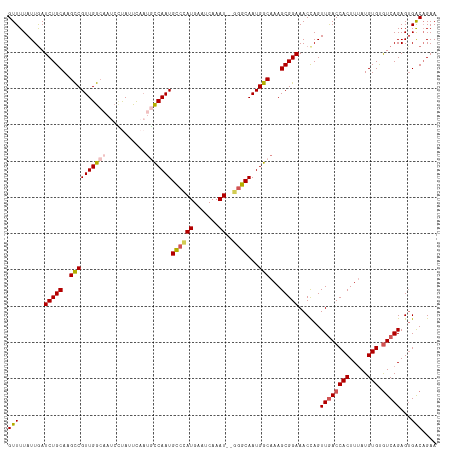
| Location | 9,538,717 – 9,538,833 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.58 |
| Shannon entropy | 0.19193 |
| G+C content | 0.47973 |
| Mean single sequence MFE | -38.68 |
| Consensus MFE | -25.80 |
| Energy contribution | -27.16 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.734355 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 9538717 116 - 22422827 CGCAGGCUCUGAACUAGUUCCGCUGAUCAUAAAGUUGGCGGUUUUAUUGAUCUGCAAGCCGUU-G-GCAAUCCUAUCCAAUGCCAAUGCCCAUGAAUCAAAU--GGGCAAUGGCAAAGCG .(((((.((.....(((..((((..((......))..))))..)))..)))))))..((.(((-(-(.........)))))((((.(((((((.......))--))))).))))...)). ( -41.70, z-score = -2.85, R) >droSim1.chrX 7590849 116 - 17042790 CGCGGGCUCUGAACUAGUUCCGCUGAUCAUAAAGUCGGCGGUUUUAUUGAUCUGCACGCCGUU-G-GCAAUCCUAUUUAAUGCCAAUGCCCAUGAAUCAAAU--GGGCAAUGGCAAAGCG (((((((.......(((..((((((((......))))))))..))).......))..(((...-)-))...)).......(((((.(((((((.......))--))))).)))))..))) ( -43.64, z-score = -3.23, R) >droSec1.super_15 302425 116 - 1954846 CGCGGGCUCUGAACUAGUUCCGCUGAUCAUAAAGUCGGCGGUUUUAUUGAUCUGCACGCCGUU-G-GCAAUCCUAUUUAAUGCCAAUGCCCAUGAAUCAAAU--GGGCAAUGGCAAAGCG (((((((.......(((..((((((((......))))))))..))).......))..(((...-)-))...)).......(((((.(((((((.......))--))))).)))))..))) ( -43.64, z-score = -3.23, R) >droEre2.scaffold_4690 13169261 112 + 18748788 CGCAGGCUCUGAACUAGUUCGGUUGAUCAUAAAGUUGGCGGUUUUAUUAAUCUGCAAGUCGUUGGCUCAGUCCUUAC------CAAUGUCUAUGAAUCAAAU--GGGCAAUGGCAAAGCG (((.(((((((((....)))))..........(((..(((..((...........))..)))..))).))))....(------((.(((((((.......))--))))).)))....))) ( -28.10, z-score = -0.57, R) >droYak2.chrX 18129206 113 - 21770863 CGCGGGCUCUGAACUAGUUCCGCUGAUCAUAAAGUCGGCGGUUUUAUUGAUCUGCAAGCCGUUGG-GCAGUCCAAUU------CAAUGCCCAUGAAUCAAAUCAAAGCAAUGGCAAAGCG .(((((.((.....(((..((((((((......))))))))..)))..)))))))..((((((((-(((........------...))))).(((......)))....))))))...... ( -36.30, z-score = -1.99, R) >consensus CGCGGGCUCUGAACUAGUUCCGCUGAUCAUAAAGUCGGCGGUUUUAUUGAUCUGCAAGCCGUU_G_GCAAUCCUAUU_AAUGCCAAUGCCCAUGAAUCAAAU__GGGCAAUGGCAAAGCG (((.((((..((..(((..((((((((......))))))))..)))....))....))))....................(((((.(((((.............))))).)))))..))) (-25.80 = -27.16 + 1.36)

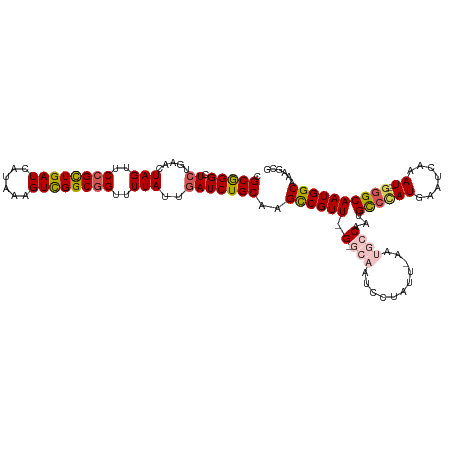

| Location | 9,538,793 – 9,538,908 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.19 |
| Shannon entropy | 0.10041 |
| G+C content | 0.44837 |
| Mean single sequence MFE | -35.68 |
| Consensus MFE | -31.18 |
| Energy contribution | -31.86 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.87 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.861818 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 9538793 115 + 22422827 CGCCAACUUUAUGAUCAGCGGAACUAGUUCAGAGCCUGCGGAUCGUAAAUUUGUGCAAUUUAUGGAGUGAUGCAA-----ACUCUGUGUUCUCACGACUUCGUGAUGAUGUGCAAUUAAA .......(((((((((.((((..((.....))..)).)).))))))))).(((..((...((((((((.......-----)))))))).(((((((....))))).))))..)))..... ( -33.20, z-score = -1.74, R) >droSim1.chrX 7590925 115 + 17042790 CGCCGACUUUAUGAUCAGCGGAACUAGUUCAGAGCCCGCGGAUCGUAAAUUUGUGCAGUUUAUGGAGUGAUGCAA-----ACUCUGUGUUCUCACGACUUCGUGAUGCUGUGCAAUUAAA .......(((((((((.((((..............)))).))))))))).(((..((((.((((((((.......-----))))))))...(((((....))))).))))..)))..... ( -37.94, z-score = -2.46, R) >droSec1.super_15 302501 113 + 1954846 CGCCGACUUUAUGAUCAGCGGAACUAGUUCAGAGCCCGCGGAUCGUAAAUUUGUGCAGUUUAUGGAGUGAUGCAA-----ACUCUGUGUUCUCACGACUUCGUGAUGCUGUGCAAUUA-- .......(((((((((.((((..............)))).))))))))).(((..((((.((((((((.......-----))))))))...(((((....))))).))))..)))...-- ( -37.94, z-score = -2.40, R) >droEre2.scaffold_4690 13169333 120 - 18748788 CGCCAACUUUAUGAUCAACCGAACUAGUUCAGAGCCUGCGAAUCGUAAAUUUGUGCAGUUUAUGGAGUGAUGUGAUGCAUACUCUGUGUUCUCACGACUUCGUGAUGCUGUGCAAUUAAA .......(((((((((....(((....)))...(....)).)))))))).(((..(((((((((((((..(((((.((((.....))))..)))))))))))))).))))..)))..... ( -31.40, z-score = -1.06, R) >droYak2.chrX 18129279 115 + 21770863 CGCCGACUUUAUGAUCAGCGGAACUAGUUCAGAGCCCGCGGAUCGUAAAUUUGUGCAGUUUAUGGAGUGAUGCAA-----ACUCUGUGUUCUCACGACUUCGUGAUGCUGUGCAAUUAAA .......(((((((((.((((..............)))).))))))))).(((..((((.((((((((.......-----))))))))...(((((....))))).))))..)))..... ( -37.94, z-score = -2.46, R) >consensus CGCCGACUUUAUGAUCAGCGGAACUAGUUCAGAGCCCGCGGAUCGUAAAUUUGUGCAGUUUAUGGAGUGAUGCAA_____ACUCUGUGUUCUCACGACUUCGUGAUGCUGUGCAAUUAAA .......(((((((((.((((..............)))).))))))))).(((..((((.((((((((............))))))))...(((((....))))).))))..)))..... (-31.18 = -31.86 + 0.68)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:29:00 2011