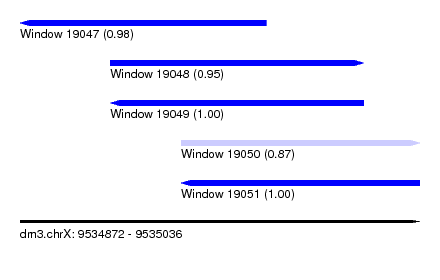
| Sequence ID | dm3.chrX |
|---|---|
| Location | 9,534,872 – 9,535,036 |
| Length | 164 |
| Max. P | 0.999476 |

| Location | 9,534,872 – 9,534,973 |
|---|---|
| Length | 101 |
| Sequences | 3 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 92.74 |
| Shannon entropy | 0.10001 |
| G+C content | 0.30363 |
| Mean single sequence MFE | -19.70 |
| Consensus MFE | -19.04 |
| Energy contribution | -19.27 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.97 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.977270 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 9534872 101 - 22422827 UUACACAGAGAGUGUGGAAAUGUGUGAGAGACGGUGUUUAUGCGUAGCACAGUUUAUGAAAUAAAAAGUUAUAGCAAAACGGAAAACAUUUUAUAUUUUAA .(((((.....)))))(((((((.....((((.(((((.......))))).)))).((..((((....))))..)).........)))))))......... ( -15.50, z-score = 0.00, R) >droSec1.super_15 298260 101 - 1954846 UUACACAGAGAGUGUGGAAGUGUGAGAGAGACGGUGUUUAUGCGUAGCACUGUUUAUGAAACAAAAAAUCAUAAAAAAUAGGAAAAUAUUUUAUAUUUUAA .(((((.....)))))(((((((((((..(((((((((.......)))))))))(((((.........)))))...............))))))))))).. ( -21.80, z-score = -3.09, R) >droSim1.chrX 7586727 101 - 17042790 UUACACAGAGAGUGUGGAAGUGUGAGAGAGACGGUGUUUAUGCGUAGCACUGUUUAUGAAACAAAAAAUCAUAAAAAAAAGGAAAAUAUUUUAUAUUUUAA .(((((.....)))))(((((((((((..(((((((((.......)))))))))(((((.........)))))...............))))))))))).. ( -21.80, z-score = -3.21, R) >consensus UUACACAGAGAGUGUGGAAGUGUGAGAGAGACGGUGUUUAUGCGUAGCACUGUUUAUGAAACAAAAAAUCAUAAAAAAAAGGAAAAUAUUUUAUAUUUUAA .(((((.....)))))(((((((((((..(((((((((.......)))))))))(((((.........)))))...............))))))))))).. (-19.04 = -19.27 + 0.23)

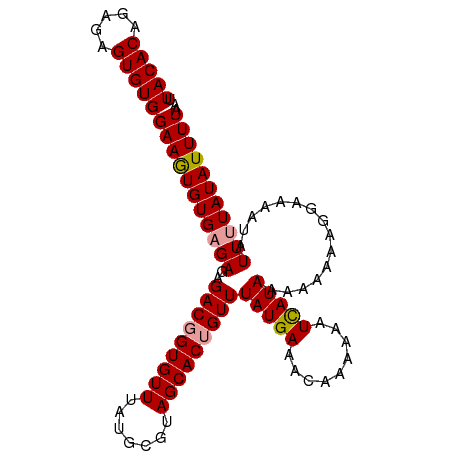
| Location | 9,534,909 – 9,535,013 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 77.91 |
| Shannon entropy | 0.41775 |
| G+C content | 0.46392 |
| Mean single sequence MFE | -27.80 |
| Consensus MFE | -14.68 |
| Energy contribution | -14.77 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.41 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.954512 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 9534909 104 + 22422827 UUAUUUCAUAAACUGUGCUACGCAUAAACACCGUCUCUCACAC----AUUUCCACACUCUCUGUGUAAAAGUGAGAGUGAAAACGCGUGGGCACAUUCCACUCUCUGC .............((((((((((............((((((..----.(((.((((.....)))).))).))))))((....)))))).))))))............. ( -26.00, z-score = -2.68, R) >dp4.chrXL_group3a 755565 103 + 2690836 CCACUGACUAAAAAGUGAGAGUGAAAAAGCGUAGGCAUAUGAC----GCACGCGAGUGUUGUGUGUGGU-GUGUGUGUGAGUACGUAUAUGUAUGAUGCACUCUUUCA .((((........))))((((((.....((.((.((((((..(----((((((((...)))))))))..-)))))).)).)).((((....))))...)))))).... ( -31.80, z-score = -1.85, R) >droEre2.scaffold_4690 13165154 104 - 18748788 UGAUUUCAUAAACAGUGCUACGCAUAAACACCGCCUCUCUCAC----ACUUCCACACUCUCUGUGUAAAAGUGAGAGUGAAAACGCGUGGGCACGUUCCACUCUCUGC ..............(((((((((..............((((((----..((.((((.....)))).))..))))))((....)))))).))))).............. ( -26.20, z-score = -1.89, R) >droYak2.chrX 18124926 108 + 21770863 CGAUUUCAUAAACAGUGCUACGCAUAAACACCGUCUCUCUCUCUCUCACUUCCACACUCUCUGCGUAAAAGUGAGAGUGAAAACGCGUGGGCACAUUCCACGCUCUGC .(.(((((......((((...)))).................(((((((((..((.(.....).))..)))))))))))))).)(((((((.....)))))))..... ( -29.30, z-score = -3.11, R) >droSec1.super_15 298297 104 + 1954846 UUGUUUCAUAAACAGUGCUACGCAUAAACACCGUCUCUCUCAC----ACUUCCACACUCUCUGUGUAAAAGUGAGAGUGAAAACGCGUGGGCACAUUCCACUCUCUGC .((((.....))))(((((((((..........((.(((((((----..((.((((.....)))).))..))))))).))....)))).))))).............. ( -26.74, z-score = -2.36, R) >droSim1.chrX 7586764 104 + 17042790 UUGUUUCAUAAACAGUGCUACGCAUAAACACCGUCUCUCUCAC----ACUUCCACACUCUCUGUGUAAAAGUGAGAGUGAAAACGCGUGGGCACAUUCCACUCUCUGC .((((.....))))(((((((((..........((.(((((((----..((.((((.....)))).))..))))))).))....)))).))))).............. ( -26.74, z-score = -2.36, R) >consensus UUAUUUCAUAAACAGUGCUACGCAUAAACACCGUCUCUCUCAC____ACUUCCACACUCUCUGUGUAAAAGUGAGAGUGAAAACGCGUGGGCACAUUCCACUCUCUGC ..............(((((((((.........(.......).............(((((((...........))))))).....)))).))))).............. (-14.68 = -14.77 + 0.09)
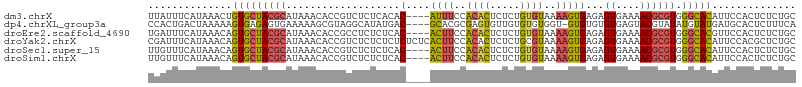
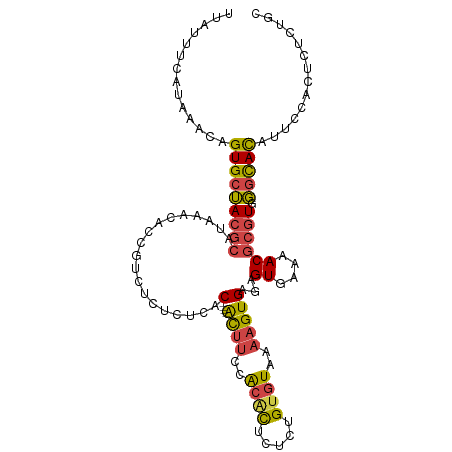
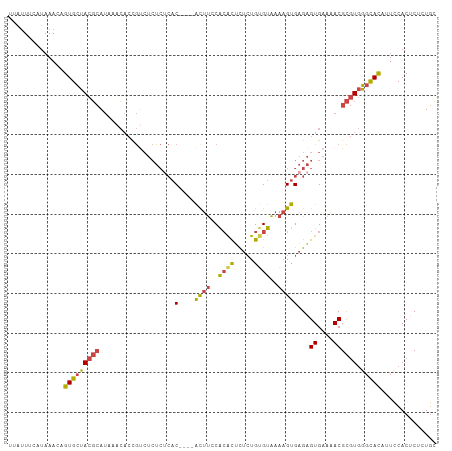
| Location | 9,534,909 – 9,535,013 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 77.91 |
| Shannon entropy | 0.41775 |
| G+C content | 0.46392 |
| Mean single sequence MFE | -36.94 |
| Consensus MFE | -25.00 |
| Energy contribution | -27.65 |
| Covariance contribution | 2.65 |
| Combinations/Pair | 1.26 |
| Mean z-score | -3.60 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.92 |
| SVM RNA-class probability | 0.999476 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 9534909 104 - 22422827 GCAGAGAGUGGAAUGUGCCCACGCGUUUUCACUCUCACUUUUACACAGAGAGUGUGGAAAU----GUGUGAGAGACGGUGUUUAUGCGUAGCACAGUUUAUGAAAUAA .......(((((.(((((..((((((...(((((((((((((((((.....))))))))..----..))))))....)))...)))))).))))).)))))....... ( -33.20, z-score = -2.36, R) >dp4.chrXL_group3a 755565 103 - 2690836 UGAAAGAGUGCAUCAUACAUAUACGUACUCACACACAC-ACCACACACAACACUCGCGUGC----GUCAUAUGCCUACGCUUUUUCACUCUCACUUUUUAGUCAGUGG ((((((((((.......(((((((((((.(........-................).))))----)).)))))....))))))))))....((((........)))). ( -19.86, z-score = -1.77, R) >droEre2.scaffold_4690 13165154 104 + 18748788 GCAGAGAGUGGAACGUGCCCACGCGUUUUCACUCUCACUUUUACACAGAGAGUGUGGAAGU----GUGAGAGAGGCGGUGUUUAUGCGUAGCACUGUUUAUGAAAUCA ...((((((((((((((....)))).))))))))))((((((((((.....))))))))))----.(((...((((((((((.......))))))))))......))) ( -40.50, z-score = -3.63, R) >droYak2.chrX 18124926 108 - 21770863 GCAGAGCGUGGAAUGUGCCCACGCGUUUUCACUCUCACUUUUACGCAGAGAGUGUGGAAGUGAGAGAGAGAGAGACGGUGUUUAUGCGUAGCACUGUUUAUGAAAUCG .....((((((.......)))))).(((((.(((((((((((((((.....))))))))))))))).)))))((((((((((.......))))))))))......... ( -47.50, z-score = -5.67, R) >droSec1.super_15 298297 104 - 1954846 GCAGAGAGUGGAAUGUGCCCACGCGUUUUCACUCUCACUUUUACACAGAGAGUGUGGAAGU----GUGAGAGAGACGGUGUUUAUGCGUAGCACUGUUUAUGAAACAA ...((((((((((((((....)))).))))))))))((((((((((.....))))))))))----((.....((((((((((.......)))))))))).....)).. ( -40.30, z-score = -4.07, R) >droSim1.chrX 7586764 104 - 17042790 GCAGAGAGUGGAAUGUGCCCACGCGUUUUCACUCUCACUUUUACACAGAGAGUGUGGAAGU----GUGAGAGAGACGGUGUUUAUGCGUAGCACUGUUUAUGAAACAA ...((((((((((((((....)))).))))))))))((((((((((.....))))))))))----((.....((((((((((.......)))))))))).....)).. ( -40.30, z-score = -4.07, R) >consensus GCAGAGAGUGGAAUGUGCCCACGCGUUUUCACUCUCACUUUUACACAGAGAGUGUGGAAGU____GUGAGAGAGACGGUGUUUAUGCGUAGCACUGUUUAUGAAAUAA ...((((((((((((((....)))).))))))))))((((((((((.....))))))))))...........((((((((((.......))))))))))......... (-25.00 = -27.65 + 2.65)

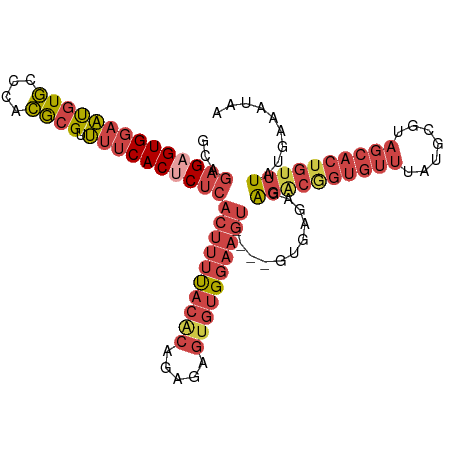
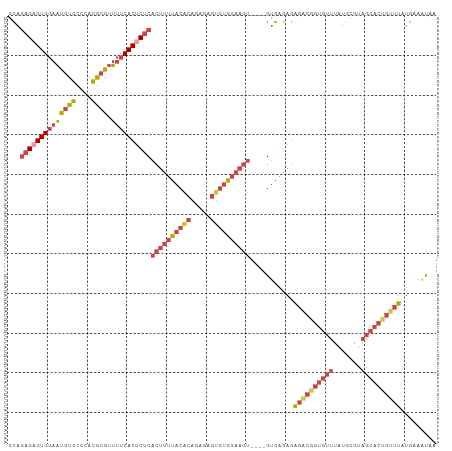
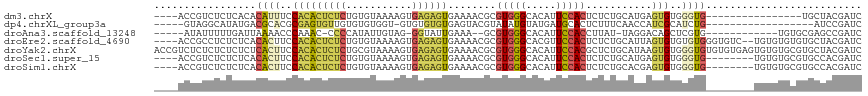
| Location | 9,534,938 – 9,535,036 |
|---|---|
| Length | 98 |
| Sequences | 7 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 65.34 |
| Shannon entropy | 0.65442 |
| G+C content | 0.50514 |
| Mean single sequence MFE | -36.04 |
| Consensus MFE | -11.35 |
| Energy contribution | -11.60 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.61 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.31 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.870553 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 9534938 98 + 22422827 ----ACCGUCUCUCACACAUUUCCACACUCUCUGUGUAAAAGUGAGAGUGAAAACGCGUGGGCACAUUCCACUCUCUGCAUGAGUGUGGGUG----------------UGCUACGAUC ----..((.(((((((...(((.((((.....)))).))).)))))))))......(((((.(((((.((((.(((.....))).)))))))----------------)))))))... ( -36.00, z-score = -3.06, R) >dp4.chrXL_group3a 755595 94 + 2690836 -----GUAGGCAUAUGACGCACGCGAGUGUUGUGUGUGGU-GUGUGUGUGAGUACGUAUAUGUAUGAUGCACUCUUUCAACCAUCGCAUCUG------------------AUCCGAUC -----.((.((((((..(((((((((...)))))))))..-)))))).)).(((((....)))))(((((...............)))))..------------------........ ( -24.76, z-score = 1.35, R) >droAna3.scaffold_13248 1329483 96 + 4840945 -----AUAUUUUUGAUUAAAACCCAAAC-CCCCAUAUUGUAG-GGUAUUGAAA--GCGUGGGCACAUUCCACCUUAU-UAGGACAGCUCGUG------------UGUGCGAGCCGAUC -----........((((....(((((((-((..........)-))).))).((--(.(((((.....))))))))..-..))...((((((.------------...)))))).)))) ( -22.40, z-score = -0.49, R) >droEre2.scaffold_4690 13165183 112 - 18748788 ----ACCGCCUCUCUCACACUUCCACACUCUCUGUGUAAAAGUGAGAGUGAAAACGCGUGGGCACGUUCCACUCUCUGCAUUAGUGUGUGUGGGUGUC--UGUGUGUGUGCUACGAUC ----...((.((.(((.(((((..((((.....))))..))))).))).))..(((((..(((((...((((.....((((....)))))))))))))--..)))))..))....... ( -36.70, z-score = -1.64, R) >droYak2.chrX 18124955 118 + 21770863 ACCGUCUCUCUCUCUCUCACUUCCACACUCUCUGCGUAAAAGUGAGAGUGAAAACGCGUGGGCACAUUCCACGCUCUGCAUAAGUGUGGGUGUGUGUGAGUGUGUGCGUGCUACGAUC ..(((.....((.(((((((((..((.(.....).))..))))))))).))....(((((.(((((((((((((((..((....))..)).))))).)))))))).))))).)))... ( -47.00, z-score = -3.97, R) >droSec1.super_15 298326 106 + 1954846 ----ACCGUCUCUCUCACACUUCCACACUCUCUGUGUAAAAGUGAGAGUGAAAACGCGUGGGCACAUUCCACUCUCUGCAUGAGUGUGGGUG--------UGUGUGCGUGCCACGAUC ----..(((.((.(((.(((((..((((.....))))..))))).))).))...((((((.((((((.((((.(((.....))).)))))))--------))).))))))..)))... ( -43.00, z-score = -3.52, R) >droSim1.chrX 7586793 106 + 17042790 ----ACCGUCUCUCUCACACUUCCACACUCUCUGUGUAAAAGUGAGAGUGAAAACGCGUGGGCACAUUCCACUCUCUGCACGAGUGUGGGUG--------UGUGUGCGUGCCACGAUC ----..(((.((.(((.(((((..((((.....))))..))))).))).))...((((((.((((((.((((.(((.....))).)))))))--------))).))))))..)))... ( -42.40, z-score = -3.11, R) >consensus ____ACCGUCUCUCUCACACUUCCACACUCUCUGUGUAAAAGUGAGAGUGAAAACGCGUGGGCACAUUCCACUCUCUGCAUGAGUGUGGGUG________UGUGUGCGUGCUACGAUC .................((((..(((((((((...........))))))........(((((.....)))))...........)))..)))).......................... (-11.35 = -11.60 + 0.25)
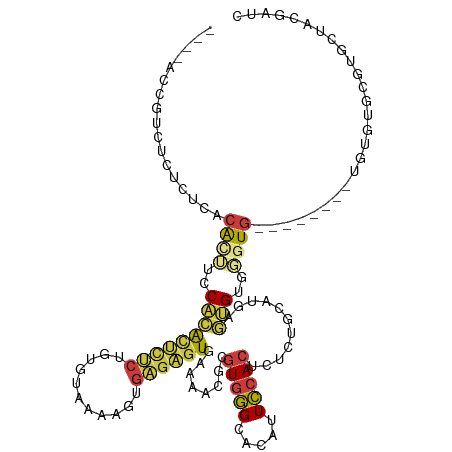
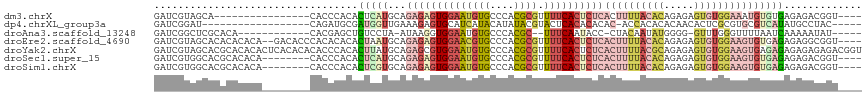
| Location | 9,534,938 – 9,535,036 |
|---|---|
| Length | 98 |
| Sequences | 7 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 65.34 |
| Shannon entropy | 0.65442 |
| G+C content | 0.50514 |
| Mean single sequence MFE | -36.50 |
| Consensus MFE | -15.45 |
| Energy contribution | -18.26 |
| Covariance contribution | 2.80 |
| Combinations/Pair | 1.48 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.28 |
| SVM RNA-class probability | 0.998176 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 9534938 98 - 22422827 GAUCGUAGCA----------------CACCCACACUCAUGCAGAGAGUGGAAUGUGCCCACGCGUUUUCACUCUCACUUUUACACAGAGAGUGUGGAAAUGUGUGAGAGACGGU---- .(((((.(((----------------((.((((.(((.....))).))))..))))).(((((((((((((.(((.((.......)).))).)))))))))))))....)))))---- ( -40.70, z-score = -4.32, R) >dp4.chrXL_group3a 755595 94 - 2690836 GAUCGGAU------------------CAGAUGCGAUGGUUGAAAGAGUGCAUCAUACAUAUACGUACUCACACACAC-ACCACACACAACACUCGCGUGCGUCAUAUGCCUAC----- ....((..------------------..((((((....((....)).))))))...(((((((((((.(........-................).)))))).)))))))...----- ( -17.06, z-score = 0.99, R) >droAna3.scaffold_13248 1329483 96 - 4840945 GAUCGGCUCGCACA------------CACGAGCUGUCCUA-AUAAGGUGGAAUGUGCCCACGC--UUUCAAUACC-CUACAAUAUGGGG-GUUUGGGUUUUAAUCAAAAAUAU----- ((((((((((....------------..))))))).((((-(.(((((((.......)))).)--))......((-(((.....)))))-..))))).....)))........----- ( -25.90, z-score = -1.14, R) >droEre2.scaffold_4690 13165183 112 + 18748788 GAUCGUAGCACACACACA--GACACCCACACACACUAAUGCAGAGAGUGGAACGUGCCCACGCGUUUUCACUCUCACUUUUACACAGAGAGUGUGGAAGUGUGAGAGAGGCGGU---- .(((((..(.(.(((((.--.....((((((...........((((((((((((((....)))).)))))))))).((((.....)))).))))))..))))).).)..)))))---- ( -39.90, z-score = -3.21, R) >droYak2.chrX 18124955 118 - 21770863 GAUCGUAGCACGCACACACUCACACACACCCACACUUAUGCAGAGCGUGGAAUGUGCCCACGCGUUUUCACUCUCACUUUUACGCAGAGAGUGUGGAAGUGAGAGAGAGAGAGACGGU .(((((..(((((.........((((...((((.(((.....))).))))..)))).....))))((((.(((((((((((((((.....))))))))))))))).)))))..))))) ( -43.24, z-score = -3.58, R) >droSec1.super_15 298326 106 - 1954846 GAUCGUGGCACGCACACA--------CACCCACACUCAUGCAGAGAGUGGAAUGUGCCCACGCGUUUUCACUCUCACUUUUACACAGAGAGUGUGGAAGUGUGAGAGAGACGGU---- ...(((((...(((((..--------...((((.(((.....))).))))..))))))))))((((((..(((.(((((((((((.....))))))))))).))).))))))..---- ( -44.00, z-score = -3.82, R) >droSim1.chrX 7586793 106 - 17042790 GAUCGUGGCACGCACACA--------CACCCACACUCGUGCAGAGAGUGGAAUGUGCCCACGCGUUUUCACUCUCACUUUUACACAGAGAGUGUGGAAGUGUGAGAGAGACGGU---- ...(((((...(((((..--------...((((.(((.....))).))))..))))))))))((((((..(((.(((((((((((.....))))))))))).))).))))))..---- ( -44.70, z-score = -3.59, R) >consensus GAUCGUAGCACGCACACA________CACCCACACUCAUGCAGAGAGUGGAAUGUGCCCACGCGUUUUCACUCUCACUUUUACACAGAGAGUGUGGAAGUGUGAGAGAGACGGU____ ..................................(((((...((((((((((((((....)))).))))))))))((((((((((.....)))))))))))))))............. (-15.45 = -18.26 + 2.80)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:28:54 2011