| Sequence ID | dm3.chrX |
|---|---|
| Location | 9,510,561 – 9,510,663 |
| Length | 102 |
| Max. P | 0.828081 |

| Location | 9,510,561 – 9,510,663 |
|---|---|
| Length | 102 |
| Sequences | 12 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 75.70 |
| Shannon entropy | 0.50396 |
| G+C content | 0.48152 |
| Mean single sequence MFE | -31.31 |
| Consensus MFE | -17.96 |
| Energy contribution | -17.67 |
| Covariance contribution | -0.29 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.828081 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
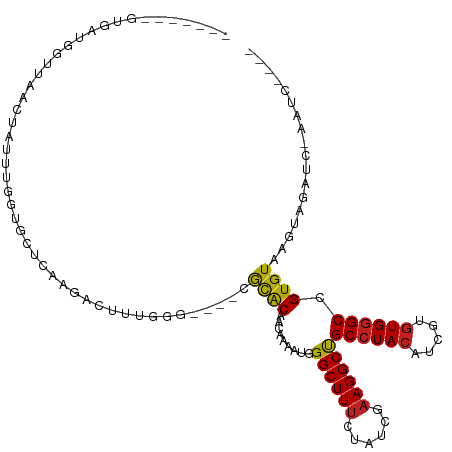
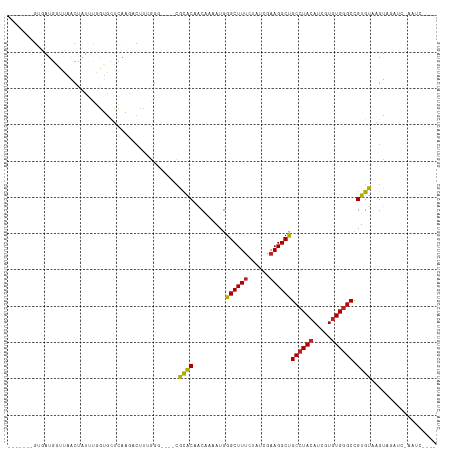
>dm3.chrX 9510561 102 + 22422827 -------GUGAUGAUUAACUAUUUGAUGCUCAAGACUUGGAG----CGCACAACAAAAUGGGCUUUCUAUCGAAGGCUGCCUACAUCGUGUGGGCCGUGUAAGUAGAUC-AAUC---- -------....(((((.(((.......((((........)))----)((((.........(((((((....)))))))((((((.....)))))).)))).))).))))-)...---- ( -33.00, z-score = -2.76, R) >droPer1.super_15 15767 110 - 2181545 GGGGAACGGAAUGGUUAACUUUUCGGUGCUUCAACCUUAUGCG---AACGCAACAAAAUGGGCUUUCUAUCGAAGGCUGCCUACAUCGUGUGGGCCGUGUGAGUACAAC-CAUC---- ((((.((.(((.((....)).))).)).))))...((((((((---..............(((((((....)))))))((((((.....))))))))))))))......-....---- ( -33.30, z-score = -1.18, R) >dp4.chrXL_group3a 730641 110 + 2690836 GGGGAACGGAAUGGUUAACUUUUCGGUGCUUCAACCUUAUGCG---AACGCAACAAAAUGGGCUUUCUAUCGAAGGCUGCCUACAUCGUGUGGGCCGUGUAAGUACAAC-CAUC---- ((((.((.(((.((....)).))).)).))))...((((((((---..............(((((((....)))))))((((((.....))))))))))))))......-....---- ( -33.20, z-score = -1.43, R) >droVir3.scaffold_12970 9338648 96 - 11907090 -----ACGGCAUGUUCAAGCAUCCAGAGGUCAAGGCUCUGGG----CGUACUACAAAAUGGGCUUUCUAUCCAAGGCCGCCUACAUUGUGUGGGCCGUGUAAGUA------------- -----(((((........((..((((((.......)))))).----.)).(((((.((((((((((......)))))).....)))).)))))))))).......------------- ( -32.30, z-score = -1.07, R) >droMoj3.scaffold_6473 12792359 104 + 16943266 -----ACGGCAUGUUCAGCCACUCCGUGCUCAAGGCUCUGGG----CGCACUACAAAAUGGGCUUUCUAUCAAAGGCCGCCUACAUCGUGUGGGCCGUGUAAGUAGCUAAACC----- -----..(((.......))).....(((((((......))))----))).((((...(((((((((......))))))((((((.....)))))))))....)))).......----- ( -34.50, z-score = -0.97, R) >droGri2.scaffold_15203 10590183 105 - 11997470 -----ACGGCAUGUUGAACCAUUCCAUGCACAAGACUUUGGC----CGCGCUACAAAAUGGGCUUCCUAUCGAAGGCUGCAUACAUCGUGUGGGCCGUGUAAGUGUUAUCAACC---- -----...(((((...........)))))..((.((((..((----.((.(((((..(((.((..(((.....)))..))...)))..))))))).))..)))).)).......---- ( -26.90, z-score = 0.80, R) >droWil1.scaffold_181096 8918845 105 - 12416693 -----ACGUUAUGUUUAACUAUUUAAUAUUUAACUAUUUGGG----CGCACAGCAAAAUGGGCUUUCUAUCCAAGGCUGCCUACAUCGUUUGGGCCGUGUAAGUUAUAUAUAUG---- -----..((((....))))......((((.(((((.....((----(.((.(((....(((((..(((.....)))..)))))....))))).))).....))))).))))...---- ( -22.30, z-score = -0.20, R) >droAna3.scaffold_13248 1302629 111 + 4840945 -------GUGAUGAUCAACUAUUUGGUUCUCAAAUCCCUGGCUUCUCGCACCGCGACAUGGGCUUUCUUUCGAAGGCUGCCUACAUUGUGUGGGCCGUGUAAGUACUUCCGAUCCAUG -------.(((.((((((....)))))).)))......(((..((.(((...)))((((((((((((....)))))))((((((.....)))))))))))..........)).))).. ( -30.80, z-score = -0.92, R) >droEre2.scaffold_4690 13143010 93 - 18748788 -------GUGAUGGUUAACUAUUUGGUGCUCAAGGCAUUGGG----CGCACAACAAAAUGGGCUUUCUAUCGAAGGCUGCCUACAUCGUGUGGGCCGUGUAAGU-------------- -------.((((((....((((((.((((((((....)))))----)))......)))))).....)))))).(.((.((((((.....)))))).)).)....-------------- ( -30.80, z-score = -1.55, R) >droYak2.chrX 18102951 102 + 21770863 -------GUGAUGGUUAACUAUUUGGUGCUCAAGGCUUUGGG----CGCACAACAAAAUGGGCUUUCUAUCGAAGGCUGCCUACAUCGUGUGGGCCGUGUAAGUAGAUC-AAUU---- -------....(((((.(((..(((((((((((....)))))----))).)))....((((((((((....)))))))((((((.....)))))))))...))).))))-)...---- ( -33.40, z-score = -2.06, R) >droSec1.super_15 276199 102 + 1954846 -------GUGAUGGUUUACUAUUUGGUGCUCAAGACUUGGGA----CGCACAACAAAAUGGGCUUUCUAUCGAAGGCUGCCUACAUCGUGUGGGCUGUGUAAGUAGAUC-AAUC---- -------....(((((((((.....((.((((.....)))))----)(((((........(((((((....)))))))((((((.....))))))))))).))))))))-)...---- ( -34.40, z-score = -3.05, R) >droSim1.chrX 7560629 102 + 17042790 -------GUGAUGGUUAACUAUUUGGUGCUCAAGACUUGGGA----CGCACAACAAAAUGGGCUUUCUAUCGAAGGCUGCCUACAUCGUGUGGGCUGUGUAAGUAGAUC-AAUC---- -------....(((((.(((.....((.((((.....)))))----)(((((........(((((((....)))))))((((((.....))))))))))).))).))))-)...---- ( -30.80, z-score = -1.96, R) >consensus _______GUGAUGGUUAACUAUUUGGUGCUCAAGACUUUGGG____CGCACAACAAAAUGGGCUUUCUAUCGAAGGCUGCCUACAUCGUGUGGGCCGUGUAAGUAGAUC_AAUC____ ...............................................((((.........((((((......))))))((((((.....)))))).)))).................. (-17.96 = -17.67 + -0.29)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:28:50 2011