| Sequence ID | dm3.chrX |
|---|---|
| Location | 9,479,254 – 9,479,357 |
| Length | 103 |
| Max. P | 0.670301 |

| Location | 9,479,254 – 9,479,357 |
|---|---|
| Length | 103 |
| Sequences | 7 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 73.21 |
| Shannon entropy | 0.46569 |
| G+C content | 0.54105 |
| Mean single sequence MFE | -35.34 |
| Consensus MFE | -16.29 |
| Energy contribution | -19.23 |
| Covariance contribution | 2.94 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.670301 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 9479254 103 - 22422827 CGUUCUUAUCGUGCCAUAAACGCGG------------AACUACAAGGGCCUGGCGGCGACGCU-CUCUCUUUUUGCUUCCGCUUAUGAGAGAGCCGCCUUUUGUCGUCGUCAGCAA .(((((...(((.......))).))------------))).......((.(((((((((((((-(((((.....((....))....))))))))........)))))))))))).. ( -38.60, z-score = -2.50, R) >droPer1.super_22 117698 93 - 1688296 --------------CAUGAACCGAG---------UUCAAUAAGGGGAGGCAGACAGCACAGCUGGACGAGCUGCUCUCCCUCUCAUGAGAGAGCCGACUGUUGUCGUCGUCAGCAA --------------..(((..((((---------(((......(((((((((.((((...))))......)))).)))))(((....)))))))((((....)))))))))).... ( -29.90, z-score = -0.12, R) >dp4.chrXL_group1e 2410260 93 + 12523060 --------------CAUGAACCGAG---------UUCAAUAAGGGGAGGCAGACAGCACAGCUGGACGAGCUGCUCUCCCUCUCAUGAGAGAGCCGACUGUUGUCGUCGUCAGCAA --------------..(((..((((---------(((......(((((((((.((((...))))......)))).)))))(((....)))))))((((....)))))))))).... ( -29.90, z-score = -0.12, R) >droSim1.chrX 7542857 102 - 17042790 CAUUCUUAUCGUGCCAUAAACGCAG------------AACUAC-AGGGCCUGGCGGCGACGCU-CUCUCUUUUUGCUUCCGCUUAUGAGAGAGCCGCCUUUUGUCGUCGUCAGCAA ..((((...(((.......))).))------------))....-...((.(((((((((((((-(((((.....((....))....))))))))........)))))))))))).. ( -35.20, z-score = -2.25, R) >droSec1.super_15 246171 102 - 1954846 CAUUCUUAUCGUGCCAUAAACGCGG------------AACUAC-AGGGCCUGGCGGCGACGCU-CUCUCUUUUUGCUUCCGCUUAUGAGAGAGCCGCCUUUUGUCGUCGUCAGCAA ........(((((.......)))))------------......-...((.(((((((((((((-(((((.....((....))....))))))))........)))))))))))).. ( -36.40, z-score = -2.26, R) >droEre2.scaffold_4690 13111598 103 + 18748788 CGUUCUUAUCACGCCAUAAACGCGG------------AACUACAAUGGCCUGGCGGCGACGCU-CUCUCUUUUUGCUUCCGCUUAUGAGAGAGCCGCGGUUUGUCGUCGUCAGCAA ............(((((........------------.......)))))((((((((((((((-(((((.....((....))....))))))))........)))))))))))... ( -36.76, z-score = -2.01, R) >droYak2.chrX 18071869 115 - 21770863 CGUUCUUAUCAUGCCAUACACGCGGAAGAACAACUACAACUACAAGGGCCUGGCGGCGACGCU-CUCUCUUUUUGCUUCCGCUUAUGAGAGAGCCGCAUUUUGUCGUCGUCAGCAA .((((((....(((.......))).))))))................((.(((((((((((((-(((((.....((....))....))))))))........)))))))))))).. ( -40.60, z-score = -3.35, R) >consensus CGUUCUUAUCGUGCCAUAAACGCGG____________AACUACAAGGGCCUGGCGGCGACGCU_CUCUCUUUUUGCUUCCGCUUAUGAGAGAGCCGCCUUUUGUCGUCGUCAGCAA ...............................................(.((((((((((((((.(((((.....((....))....))))))))........)))))))))))).. (-16.29 = -19.23 + 2.94)

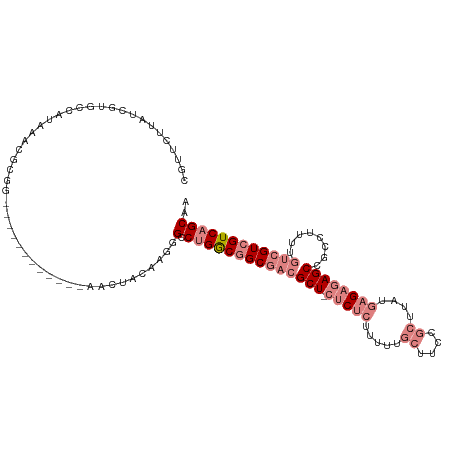

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:28:43 2011