| Sequence ID | dm3.chrX |
|---|---|
| Location | 9,478,679 – 9,478,792 |
| Length | 113 |
| Max. P | 0.908032 |

| Location | 9,478,679 – 9,478,792 |
|---|---|
| Length | 113 |
| Sequences | 7 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 63.37 |
| Shannon entropy | 0.66969 |
| G+C content | 0.48648 |
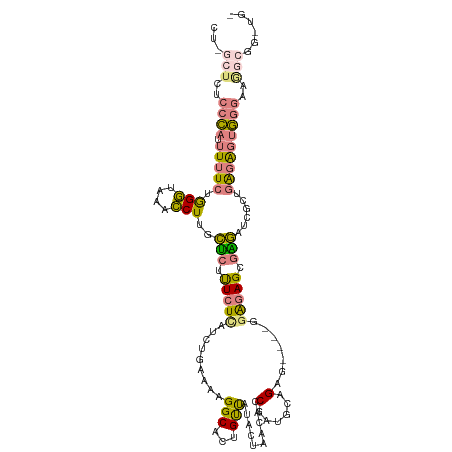
| Mean single sequence MFE | -38.75 |
| Consensus MFE | -9.00 |
| Energy contribution | -9.82 |
| Covariance contribution | 0.82 |
| Combinations/Pair | 1.46 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.23 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.908032 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
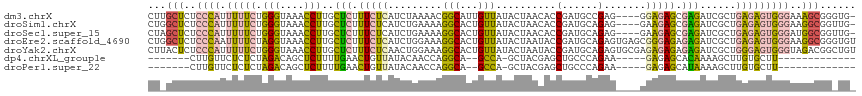
>dm3.chrX 9478679 113 + 22422827 CUUGCUCUCCCAUUUUUCUGGGUAAACCUUGCUCUUUCUCAUCUAAAACGGCAUUGUUAUACUAACACCGAUGCCGAG----GGAGAGCGAGAUCGCUGAGAGUGGGAAAGCGGGUG- ((((((.((((((...(((.(((....((((((((..(((.........(((((((............))))))))))----..))))))))...))).))))))))).))))))..- ( -47.70, z-score = -4.97, R) >droSim1.chrX 7542274 113 + 17042790 CUGGCUCUCCCAUUUUUCUGGGUAAACCUUGCUCUUUCUCAUCUGAAAAGGCACUGUUAUACUAACACCGAUGCAGAG----GAAGAGCGAGAUCGCUGAGAGUGGGAAGGCGGUUG- ...((..((((((...(((.(((....(((((((((((((..((....))(((.(((((...)))))....))).)))----))))))))))...))).)))))))))..)).....- ( -44.20, z-score = -3.49, R) >droSec1.super_15 245590 113 + 1954846 CUAGCUCUCCCAUUUUUCUGGGUAAACCUUGCUCUUUCUCAUCUGAAAAGGCACUGUUAUACUAACACCGAUGCAGAG----GAAGAGCGAGAUCGCUGAGAGUGGGAUGGCGGUUG- ...(((.((((((...(((.(((....(((((((((((((..((....))(((.(((((...)))))....))).)))----))))))))))...))).))))))))).))).....- ( -44.00, z-score = -3.76, R) >droEre2.scaffold_4690 13110986 118 - 18748788 CUGGCUCUCCCAAUUUUCUAGGUAAACCUUGCUCUUUCUCAUCUGGAAAGGCACUGUUAUACUAAUACCGAUGCAGAGUGAGCGGGAGAGAGAUCGCUGAGAGUGGGAAGGCGGGUGU ((.((..(((((.(((((.(((....))).((((((((((.(((....)))..(((((.((((.............)))))))))))))))))..)).))))))))))..)).))... ( -37.52, z-score = -0.45, R) >droYak2.chrX 18071259 118 + 21770863 CUUACUCUCCCAUUUUUCUGGGUAAACCUUGCUCUUUCUCAACUGGAAAGGCACUGUUAUACUAAUACCGAUGCAGAGUGCGAGAGAGAGAGAUCGCUGGGAGUGGGUAGACGGCUGU .(((((((((((....((((((....)))..(((((((((..((....))(((((.((((..........))).).)))))))))))))))))....))))))..)))))........ ( -36.40, z-score = -0.87, R) >dp4.chrXL_group1e 2409884 90 - 12523060 -------CUUGUUCUCUCUAGACAGCUCUUUUGAACUGUUAUACAACCAGGCA--GCCA-GCUACGAGCUGCCCAGAA-----GAGAGCACAAAAGCUUGUGCUU------------- -------.((((.((((((.(((((.((....)).))))).........((((--(((.-.....).))))))....)-----))))).))))((((....))))------------- ( -30.90, z-score = -2.44, R) >droPer1.super_22 117323 90 + 1688296 -------CUUGUUCUCUCUAGACAGCUCUUUUGAACUGUUAUACAACCAGGCA--GCCA-GCUACGAGCUGCCCAGAA-----GAGAGCAUAAAAGCUUGUGCUU------------- -------..((((((((((.(((((.((....)).))))).........((((--(((.-.....).)))))).)).)-----)))))))...((((....))))------------- ( -30.50, z-score = -2.45, R) >consensus CU_GCUCUCCCAUUUUUCUGGGUAAACCUUGCUCUUUCUCAUCUGAAAAGGCACUGUUAUACUAACACCGAUGCAGAG____GGAGAGCGAGAUCGCUGAGAGUGGGAAGGCGG_UG_ ((.((((((..........(((....)))..(((.(((((..........(((.((............)).))).........))))).)))......)))))).))........... ( -9.00 = -9.82 + 0.82)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:28:42 2011