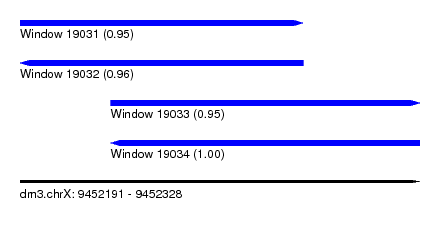
| Sequence ID | dm3.chrX |
|---|---|
| Location | 9,452,191 – 9,452,328 |
| Length | 137 |
| Max. P | 0.995205 |

| Location | 9,452,191 – 9,452,288 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 64.48 |
| Shannon entropy | 0.56361 |
| G+C content | 0.47020 |
| Mean single sequence MFE | -25.88 |
| Consensus MFE | -16.06 |
| Energy contribution | -19.00 |
| Covariance contribution | 2.94 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.947479 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 9452191 97 + 22422827 -----UGAGUAUUAUUGUCGAAUCAUUAUCAAAGUCGUUUCCUGGUUUGGAUUCCUCGGAUUUGGUUGAGGAACCAAGCCAUGUGCCGACAUUUCGUCGGCA -----.....................................((((((((.((((((((......))))))))))))))))..(((((((.....))))))) ( -32.70, z-score = -2.89, R) >droSim1.chrX 9470300 96 - 17042790 -----UGAGUAAUAUUAUCGAAUCAUUAUCAAAUUCGUUUCCUAGUUUGGAUUCCUCGGAAUCGGGCGAGGACCCAAGCC-UGUGCCGACAUUUCGUCGGCA -----.............(((((.........))))).......((((((..((((((........)))))).)))))).-..(((((((.....))))))) ( -28.50, z-score = -1.57, R) >droSec1.super_15 218224 96 + 1954846 -----UGAGUAACAUUAUCGAAUCAUUAUCAAAUUCGUUUCCUGGUUUGGAUUCCUCGGAUUCGGGCGACGACCCAAGCC-UGUGCCGACAUUUCGUCGGCA -----......(((....(((((.........)))))......(((((((..((((((....)))).))....)))))))-)))((((((.....)))))). ( -26.70, z-score = -0.72, R) >droEre2.scaffold_4784 23198909 90 - 25762168 CGAAUUAAAUCCCCUCCACUGGCAAAUCCACGCCUCGUCAUCUGGAAAAGAUUCCUCCCUCUCAAGCAAGGAAUUAGGCU-CAUCGCGACA----------- ....................(((........)))..(((..........(((((((..((....))..)))))))..((.-....))))).----------- ( -15.60, z-score = 0.14, R) >consensus _____UGAGUAACAUUAUCGAAUCAUUAUCAAAUUCGUUUCCUGGUUUGGAUUCCUCGGAUUCGGGCGAGGAACCAAGCC_UGUGCCGACAUUUCGUCGGCA ...........................................(((((((.(((((((........))))))))))))))....((((((.....)))))). (-16.06 = -19.00 + 2.94)



| Location | 9,452,191 – 9,452,288 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 64.48 |
| Shannon entropy | 0.56361 |
| G+C content | 0.47020 |
| Mean single sequence MFE | -28.45 |
| Consensus MFE | -13.34 |
| Energy contribution | -16.40 |
| Covariance contribution | 3.06 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.959849 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 9452191 97 - 22422827 UGCCGACGAAAUGUCGGCACAUGGCUUGGUUCCUCAACCAAAUCCGAGGAAUCCAAACCAGGAAACGACUUUGAUAAUGAUUCGACAAUAAUACUCA----- (((((((.....)))))))..(((.((((((((((..........)))))).)))).)))(....)....((((.......))))............----- ( -29.20, z-score = -3.49, R) >droSim1.chrX 9470300 96 + 17042790 UGCCGACGAAAUGUCGGCACA-GGCUUGGGUCCUCGCCCGAUUCCGAGGAAUCCAAACUAGGAAACGAAUUUGAUAAUGAUUCGAUAAUAUUACUCA----- (((((((.....)))))))(.-((.(((((((((((........))))).)))))).)).)....((((((.......)))))).............----- ( -30.70, z-score = -2.75, R) >droSec1.super_15 218224 96 - 1954846 UGCCGACGAAAUGUCGGCACA-GGCUUGGGUCGUCGCCCGAAUCCGAGGAAUCCAAACCAGGAAACGAAUUUGAUAAUGAUUCGAUAAUGUUACUCA----- (((((((.....)))))))..-((.((((((....))))))..))(((...(((......)))..((((((.......)))))).........))).----- ( -30.50, z-score = -2.01, R) >droEre2.scaffold_4784 23198909 90 + 25762168 -----------UGUCGCGAUG-AGCCUAAUUCCUUGCUUGAGAGGGAGGAAUCUUUUCCAGAUGACGAGGCGUGGAUUUGCCAGUGGAGGGGAUUUAAUUCG -----------....((....-.))....((((((......)))))).(((((((((((((....)..((((......))))..))))))))))))...... ( -23.40, z-score = 0.58, R) >consensus UGCCGACGAAAUGUCGGCACA_GGCUUGGGUCCUCGCCCGAAUCCGAGGAAUCCAAACCAGGAAACGAAUUUGAUAAUGAUUCGAUAAUAUUACUCA_____ (((((((.....)))))))...((.(((((((((((........))))).)))))).)).(....)....((((.......))))................. (-13.34 = -16.40 + 3.06)


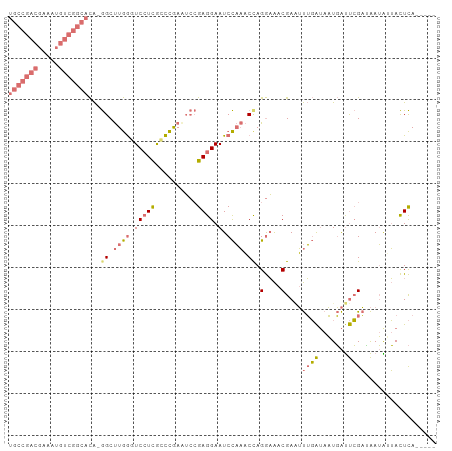
| Location | 9,452,222 – 9,452,328 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 70.77 |
| Shannon entropy | 0.47399 |
| G+C content | 0.50274 |
| Mean single sequence MFE | -34.38 |
| Consensus MFE | -20.02 |
| Energy contribution | -23.90 |
| Covariance contribution | 3.88 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.949962 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 9452222 106 + 22422827 GUUUCCUGGUUUGGAUUCCUCGGAUUUGGUUGAGGAACCAAGCCAUGUGCCGACAUUUCGUCGGCAAUGUAGCUGUAACGAUGUGUACGCGCUUUGAGGUGACUGU (((.((((((((((.((((((((......)))))))))))))))..((((..((((.((((((((......))))..)))).))))..))))....))).)))... ( -42.70, z-score = -3.86, R) >droSim1.chrX 9470331 105 - 17042790 GUUUCCUAGUUUGGAUUCCUCGGAAUCGGGCGAGGACCCAAGCC-UGUGCCGACAUUUCGUCGGCACUGUAGCUGUAACGAUGUGUACGCGCUUUGAGGUGACUGU (((.(((.((((((..((((((........)))))).)))))).-.((((((((.....))))))))...........(((.(((....))).)))))).)))... ( -39.20, z-score = -2.00, R) >droSec1.super_15 218255 105 + 1954846 GUUUCCUGGUUUGGAUUCCUCGGAUUCGGGCGACGACCCAAGCC-UGUGCCGACAUUUCGUCGGCACUGUAGCUGUAACGAUGUGUACGCGCUUUGAGGUGACUGU (((.((((((((((..((((((....)))).))....)))))))-.((((((((.....))))))))...........(((.(((....))).)))))).)))... ( -37.10, z-score = -1.25, R) >droEre2.scaffold_4784 23198945 89 - 25762168 GUCAUCUGGAAAAGAUUCCUCCCUCUCAAGCAAGGAAUUAGGCU---------CAUCGC---GACAGUGUUGCUGUAUUUAAGAGCAAAUGUCUUAAA-UGU---- ..(((..(((...(((((((..((....))..)))))))..(((---------(.....---.((((.....))))......)))).....)))...)-)).---- ( -18.50, z-score = -0.30, R) >consensus GUUUCCUGGUUUGGAUUCCUCGGAUUCGGGCGAGGAACCAAGCC_UGUGCCGACAUUUCGUCGGCACUGUAGCUGUAACGAUGUGUACGCGCUUUGAGGUGACUGU (((.((((((((((.(((((((........))))))))))))))..((((((((.....))))))))...........(((.(((....))).)))))).)))... (-20.02 = -23.90 + 3.88)

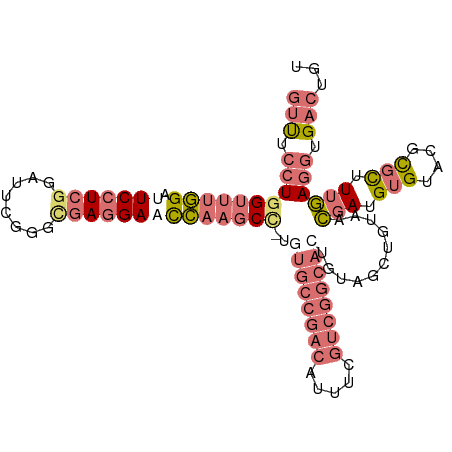
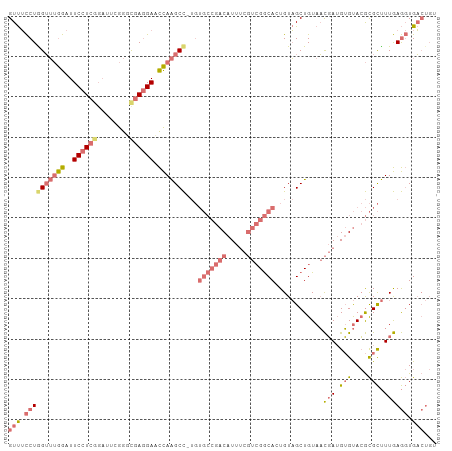
| Location | 9,452,222 – 9,452,328 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 70.77 |
| Shannon entropy | 0.47399 |
| G+C content | 0.50274 |
| Mean single sequence MFE | -31.28 |
| Consensus MFE | -18.11 |
| Energy contribution | -19.55 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.78 |
| SVM RNA-class probability | 0.995205 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 9452222 106 - 22422827 ACAGUCACCUCAAAGCGCGUACACAUCGUUACAGCUACAUUGCCGACGAAAUGUCGGCACAUGGCUUGGUUCCUCAACCAAAUCCGAGGAAUCCAAACCAGGAAAC .......((....(((..(((........))).)))....(((((((.....)))))))..(((.((((((((((..........)))))).)))).))))).... ( -32.00, z-score = -3.30, R) >droSim1.chrX 9470331 105 + 17042790 ACAGUCACCUCAAAGCGCGUACACAUCGUUACAGCUACAGUGCCGACGAAAUGUCGGCACA-GGCUUGGGUCCUCGCCCGAUUCCGAGGAAUCCAAACUAGGAAAC .......((((..(((..(((........))).)))...((((((((.....)))))))).-((.((((((....))))))..))))))..(((......)))... ( -36.20, z-score = -2.97, R) >droSec1.super_15 218255 105 - 1954846 ACAGUCACCUCAAAGCGCGUACACAUCGUUACAGCUACAGUGCCGACGAAAUGUCGGCACA-GGCUUGGGUCGUCGCCCGAAUCCGAGGAAUCCAAACCAGGAAAC .......((((..(((..(((........))).)))...((((((((.....)))))))).-((.((((((....))))))..))))))..(((......)))... ( -36.20, z-score = -2.78, R) >droEre2.scaffold_4784 23198945 89 + 25762168 ----ACA-UUUAAGACAUUUGCUCUUAAAUACAGCAACACUGUC---GCGAUG---------AGCCUAAUUCCUUGCUUGAGAGGGAGGAAUCUUUUCCAGAUGAC ----.((-(((.......((.(((((((..((((.....)))).---((((.(---------((.....))).))))))))))).))((((....))))))))).. ( -20.70, z-score = -1.29, R) >consensus ACAGUCACCUCAAAGCGCGUACACAUCGUUACAGCUACAGUGCCGACGAAAUGUCGGCACA_GGCUUGGGUCCUCGCCCGAAUCCGAGGAAUCCAAACCAGGAAAC .......((((......................(((...((((((((.....))))))))..)))((((((....))))))....))))..(((......)))... (-18.11 = -19.55 + 1.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:28:40 2011