| Sequence ID | dm3.chrX |
|---|---|
| Location | 9,442,262 – 9,442,333 |
| Length | 71 |
| Max. P | 0.557644 |

| Location | 9,442,262 – 9,442,333 |
|---|---|
| Length | 71 |
| Sequences | 13 |
| Columns | 78 |
| Reading direction | reverse |
| Mean pairwise identity | 68.07 |
| Shannon entropy | 0.71347 |
| G+C content | 0.32767 |
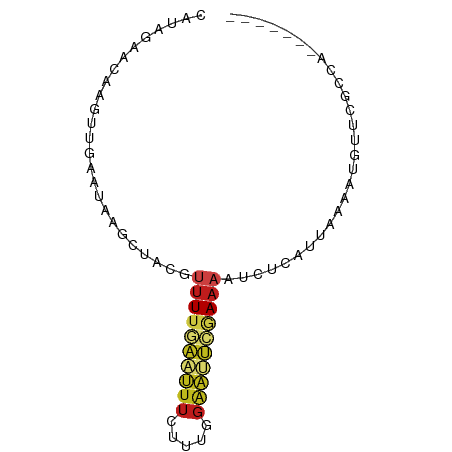
| Mean single sequence MFE | -12.02 |
| Consensus MFE | -4.75 |
| Energy contribution | -5.08 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.60 |
| Mean z-score | -0.88 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.557644 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
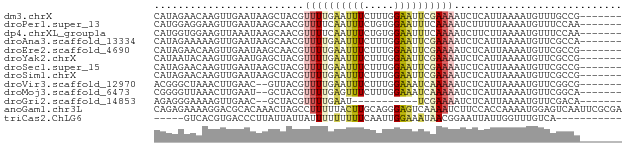
>dm3.chrX 9442262 71 - 22422827 CAUAGAACAAGUUGAAUAAGCUACGUUUUGAAUUUCUUUGGAAUUCGAAAAUCUCAUUAAAAUGUUUGCCG------- ....(((((((((.....))))...((((((((((.....))))))))))............)))))....------- ( -12.20, z-score = -1.14, R) >droPer1.super_13 110419 71 + 2293547 CAUGGAGGAAGUUGAAUAAGCAACGUUUUCAAUUUCUGUGGAAUUUCAAAAUCUUUUUAAAAUGUUUCCAA------- ..((((((((((((((...........)))))))))).(((....)))..................)))).------- ( -11.80, z-score = -0.42, R) >dp4.chrXL_group1a 1614464 71 + 9151740 CAUGGUGGAAGUUAAAUAAGCAACGUUUUCAAUUUCUGUGGAAUUUCAAAAUCUUCUUAAAAUGUUUCCAA------- .....((((((.....((((.((.(((((.((.(((....))).)).))))).)))))).....)))))).------- ( -9.60, z-score = -0.38, R) >droAna3.scaffold_13334 1491479 71 + 1562580 CAUAGAAAAAGUUGAAUAAGCAACGUUUUGAAUUUCUUUGGAAUUCGAAAAUCUCAUUAAAAUGUUCGCCA------- ....(((...((((......)))).((((((((((.....))))))))))..............)))....------- ( -11.80, z-score = -0.89, R) >droEre2.scaffold_4690 13075516 71 + 18748788 CAUAGAACAAGUUGAAUAAGCAACGUUUUGAAUUUCUUUGGAAUUCGAAAAUCUCAUUAAAAUGUUCGCCG------- ....(((((.((((......)))).((((((((((.....))))))))))............)))))....------- ( -16.20, z-score = -2.60, R) >droYak2.chrX 18036272 71 - 21770863 CAUAAUACAAGUUGAAUGAGCUACGUUUUGAAUUUCUUUGGAAUUCGAAAAUCUCAUUAAAAUGUUCGCCG------- ......(((..((((.((((.....((((((((((.....))))))))))..))))))))..)))......------- ( -13.60, z-score = -1.62, R) >droSec1.super_15 209110 71 - 1954846 CAUAGAACAAGUUGAAUAAGCUACGUUUUGAAUUUCUUUGGAAUUCGAAAAUCUCAUUAAAAUGUUCGCCG------- ....(((((((((.....))))...((((((((((.....))))))))))............)))))....------- ( -14.70, z-score = -2.07, R) >droSim1.chrX 7519598 71 - 17042790 CAUAGAACAAGUUGAAUAAGCUACGUUUUGAAUUUCUUUGGAAUUCGAAAAUCUCAUUAAAAUGUUCGCCG------- ....(((((((((.....))))...((((((((((.....))))))))))............)))))....------- ( -14.70, z-score = -2.07, R) >droVir3.scaffold_12970 6095076 69 - 11907090 ACGGGCUAAACUUGAAC--GUUACGUUUUGAAUUUCUUUGGAAAUCAAAAAUCUCAUUAAAAUGUUCGGCG------- ....((..(((......--......((((((.((((....)))))))))).............)))..)).------- ( -11.31, z-score = -0.83, R) >droMoj3.scaffold_6473 1421358 69 + 16943266 CGGGGUUAAACUUGAAU--GCUACGUUUUGAGUUUCUUUGGAAAUCAAAAAUCUCAUUAAAAUGUUCGGCA------- ....((..(((...(((--(.....((((((.((((....))))))))))....)))).....)))..)).------- ( -12.10, z-score = -0.92, R) >droGri2.scaffold_14853 943025 58 + 10151454 AGAGGGAAAAGUUGAAC--GCUACGUUUUGAAU-----------UCGAAAAUCUCAUUAAAAUGUUCGACA------- ..........(((((((--(.....(((((...-----------.)))))............)))))))).------- ( -8.33, z-score = 0.39, R) >anoGam1.chr3L 27040935 78 - 41284009 CAGAGAAAAGGACGCACAAACUAGCCUUUUUACUUGCAGGGAGUCAAAAUCUUCCACCAAAAUGGAGUCAAUUCGCGA .((.(((((((.(..........)))))))).))(((.(((((.......))))).((.....)).........))). ( -15.10, z-score = -0.35, R) >triCas2.ChLG6 6802173 62 - 13544221 -----GUCACGUGACCCUUAUUAUUAUUUUUUUUUCAAUUGGAAAUAACGGAAUUAUUGGUUUGUCA----------- -----(.((...((((..(((..((((((((.........))))))))..)...))..)))))).).----------- ( -4.80, z-score = 1.45, R) >consensus CAUAGAACAAGUUGAAUAAGCUACGUUUUGAAUUUCUUUGGAAUUCGAAAAUCUCAUUAAAAUGUUCGCCA_______ .........................((((((((((.....))))))))))............................ ( -4.75 = -5.08 + 0.34)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:28:35 2011