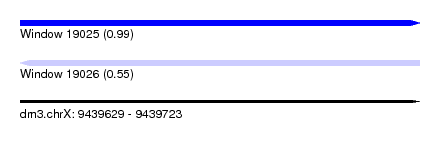
| Sequence ID | dm3.chrX |
|---|---|
| Location | 9,439,629 – 9,439,723 |
| Length | 94 |
| Max. P | 0.988730 |

| Location | 9,439,629 – 9,439,723 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 73.62 |
| Shannon entropy | 0.45314 |
| G+C content | 0.47242 |
| Mean single sequence MFE | -22.23 |
| Consensus MFE | -9.72 |
| Energy contribution | -11.58 |
| Covariance contribution | 1.86 |
| Combinations/Pair | 1.18 |
| Mean z-score | -3.00 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.33 |
| SVM RNA-class probability | 0.988730 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
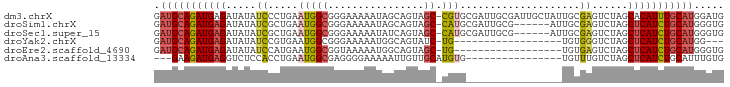
>dm3.chrX 9439629 94 + 22422827 CAUCCAUGCAAAUGUGCUAGACUCGCAAUAGCAAUCGCAAUCGCACG-GCUACUGCUAUUUUUCCCGCCAUUCAGGGAUAUAUCUCAUCUGCAUC .....(((((.(((.(.((.....(((.((((....((....))...-)))).)))......((((........))))..)).).))).))))). ( -22.30, z-score = -1.42, R) >droSim1.chrX 7517191 88 + 17042790 CACCCAUGCAGAUGAGCUAGACUCGCAAU------CGCAAUCGCAUG-GCUACUGCUAUUUUUCCCGCCAUUCAGCGAUAUAUCUCAUCUGCAUC .....(((((((((((.((...((((...------.((....))(((-((................)))))...))))..)).))))))))))). ( -29.39, z-score = -4.23, R) >droSec1.super_15 206547 88 + 1954846 CACCCAUGCAGAUGAGCUAGACUCGCAAU------CGCAAUCGCAUG-GCUACUGAUAUUUUUCCCGCCAUUCAGCGAUAUAUCUCAUCUGCAUC .....(((((((((((.((...((((...------.((....))(((-((....((......))..)))))...))))..)).))))))))))). ( -30.10, z-score = -4.52, R) >droYak2.chrX 18032967 73 + 21770863 ---CCAUGCAGAUGAGCUAGACCCACA------------------CA-GAUACUGCCAUUUUUCCCGCCAUUCACGGAUAUAUCUCAUCUGCAUC ---..(((((((((((.((........------------------((-(...))).........(((.......)))...)).))))))))))). ( -18.70, z-score = -3.62, R) >droEre2.scaffold_4690 13073019 76 - 18748788 CACCCAUGCAGAUGAGCUAGACUCACA------------------CA-GCUACUGCCAUUUUUACCGCCAUUCAUGGAUAUAUCUCAUCUGCAUC .....(((((((((((.((........------------------..-((....))...........(((....)))...)).))))))))))). ( -19.20, z-score = -3.25, R) >droAna3.scaffold_13334 1488798 76 - 1562580 CACAAAUGCAGAUGAGCUAGACAAACA----------------CACAUGCAACAAUUUUUCCCCUCGCCAUUCAGGUGGAGACCUCAUCUUC--- .........(((((((...(.((....----------------....))).......((((((((........))).))))).)))))))..--- ( -13.70, z-score = -0.94, R) >consensus CACCCAUGCAGAUGAGCUAGACUCACA________________CACA_GCUACUGCUAUUUUUCCCGCCAUUCAGGGAUAUAUCUCAUCUGCAUC .....(((((((((((................................((....))......((((........)))).....))))))))))). ( -9.72 = -11.58 + 1.86)

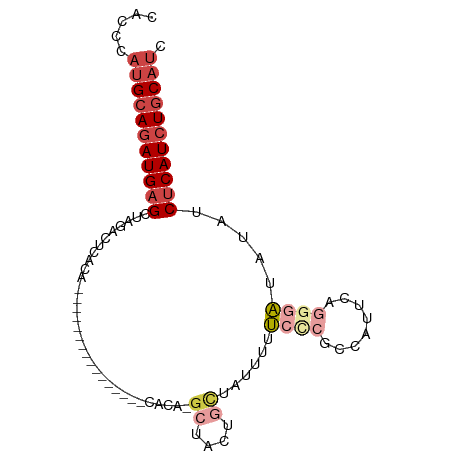
| Location | 9,439,629 – 9,439,723 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 73.62 |
| Shannon entropy | 0.45314 |
| G+C content | 0.47242 |
| Mean single sequence MFE | -26.57 |
| Consensus MFE | -11.65 |
| Energy contribution | -11.98 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.545321 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 9439629 94 - 22422827 GAUGCAGAUGAGAUAUAUCCCUGAAUGGCGGGAAAAAUAGCAGUAGC-CGUGCGAUUGCGAUUGCUAUUGCGAGUCUAGCACAUUUGCAUGGAUG .(((((((((((((...((((........))))......((((((((-..(((....)))...))))))))..))))....)))))))))..... ( -32.10, z-score = -2.65, R) >droSim1.chrX 7517191 88 - 17042790 GAUGCAGAUGAGAUAUAUCGCUGAAUGGCGGGAAAAAUAGCAGUAGC-CAUGCGAUUGCG------AUUGCGAGUCUAGCUCAUCUGCAUGGGUG .(((((((((((.....((((...(((((................))-)))((....)).------...))))......)))))))))))..... ( -30.89, z-score = -2.04, R) >droSec1.super_15 206547 88 - 1954846 GAUGCAGAUGAGAUAUAUCGCUGAAUGGCGGGAAAAAUAUCAGUAGC-CAUGCGAUUGCG------AUUGCGAGUCUAGCUCAUCUGCAUGGGUG .(((((((((((.....((((...(((((..((......))....))-)))((....)).------...))))......)))))))))))..... ( -32.20, z-score = -2.62, R) >droYak2.chrX 18032967 73 - 21770863 GAUGCAGAUGAGAUAUAUCCGUGAAUGGCGGGAAAAAUGGCAGUAUC-UG------------------UGUGGGUCUAGCUCAUCUGCAUGG--- .(((((((((((.....(((((.....))))).......((((...)-))------------------)..........)))))))))))..--- ( -24.80, z-score = -2.58, R) >droEre2.scaffold_4690 13073019 76 + 18748788 GAUGCAGAUGAGAUAUAUCCAUGAAUGGCGGUAAAAAUGGCAGUAGC-UG------------------UGUGAGUCUAGCUCAUCUGCAUGGGUG .(((((((((((.((.(..(((..(((((.((.......))....))-))------------------))))..).)).)))))))))))..... ( -23.00, z-score = -1.54, R) >droAna3.scaffold_13334 1488798 76 + 1562580 ---GAAGAUGAGGUCUCCACCUGAAUGGCGAGGGGAAAAAUUGUUGCAUGUG----------------UGUUUGUCUAGCUCAUCUGCAUUUGUG ---(.(((((((.(((((.((.....))...))))).........(((....----------------....)))....))))))).)....... ( -16.40, z-score = 0.48, R) >consensus GAUGCAGAUGAGAUAUAUCCCUGAAUGGCGGGAAAAAUAGCAGUAGC_CGUG________________UGCGAGUCUAGCUCAUCUGCAUGGGUG .(((((((((((..........((.(.(((......................................))).).))...)))))))))))..... (-11.65 = -11.98 + 0.33)
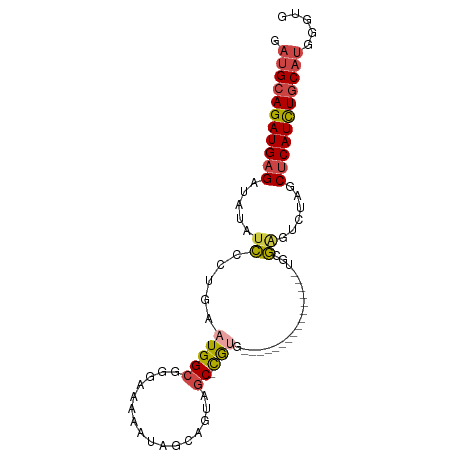
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:28:34 2011