| Sequence ID | dm3.chrX |
|---|---|
| Location | 9,426,368 – 9,426,459 |
| Length | 91 |
| Max. P | 0.508504 |

| Location | 9,426,368 – 9,426,459 |
|---|---|
| Length | 91 |
| Sequences | 10 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 79.20 |
| Shannon entropy | 0.39862 |
| G+C content | 0.44100 |
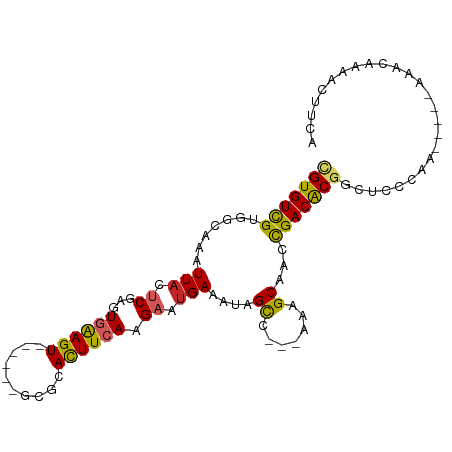
| Mean single sequence MFE | -20.86 |
| Consensus MFE | -13.03 |
| Energy contribution | -12.62 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.508504 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
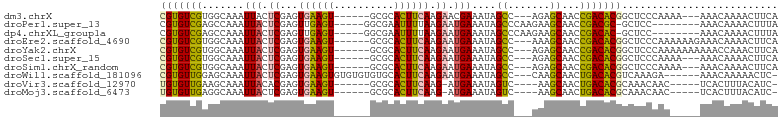
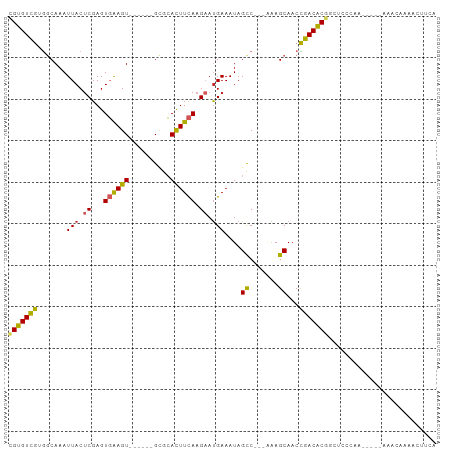
>dm3.chrX 9426368 91 - 22422827 CGUGUCGUGGCAAAUUACUCGAGUGAAGU------GCGCACUUCAAGAACGAAAUAGCC---AGAGCAACCGACACGGCUCCCAAAA---AAACAAAACUUCA (((((((((((.......(((..((((((------....))))))....)))....)))---).......)))))))..........---............. ( -21.41, z-score = -1.20, R) >droPer1.super_13 97008 89 + 2293547 CGUGUCGAGCCAAAUUACUCGAGUUGAGU-----GGCGAAUUUUAAGAAUGAAAUAGCCCAAGAAGCAACCGACGC-GCUCC--------AAACAAAACUUUA (((((((.((......((((.....))))-----(((..((((((....)))))).)))......))...))))))-)....--------............. ( -19.00, z-score = -1.18, R) >dp4.chrXL_group1a 1600795 89 + 9151740 CGUGUCGAGCCAAAUUACUCGAGUUGAGU-----GGCGAAUUUUAAGAAUGAAAUAGCCCAAGAAGCAACCGACAC-GCUCC--------AAACAAAACUUUA (((((((.((......((((.....))))-----(((..((((((....)))))).)))......))...))))))-)....--------............. ( -19.40, z-score = -1.85, R) >droEre2.scaffold_4690 13059219 94 + 18748788 CGUGUCGUGGCAAAUUACUCGAGUGAAGU------GCGCACUUCAAGAAUGAAAUAGCC---AAAGCAACCGACACGGCUCCCAAAAAAGAAACAAAACUUCA (((((((((((...(((.((...((((((------....)))))).)).)))....)))---).......))))))).......................... ( -22.41, z-score = -1.73, R) >droYak2.chrX 18019056 94 - 21770863 CGUGUCGUGGCAAAUUACUCGAGUGAAGU------GCGCACUUCAAGAAUGAAAUAGCC---AGAGCAACCGACACGGCUCCCAAAAAAAAAACCAAACUUCA (((((((((((...(((.((...((((((------....)))))).)).)))....)))---).......))))))).......................... ( -22.41, z-score = -1.70, R) >droSec1.super_15 190408 91 - 1954846 CGUGUCGUGGCAAAUUACUCGAGUGAAGU------GCGCACUUCAAGAAUGAAAUAGCC---AGAGCAACCGACACGGCUCCCAAAA---AAACAAAACUUCA (((((((((((...(((.((...((((((------....)))))).)).)))....)))---).......)))))))..........---............. ( -22.41, z-score = -1.59, R) >droSim1.chrX_random 2710347 91 - 5698898 CGUGUCGUGGCAAAUUACUCGAGUGAAGU------GCGCACUUCAAGAAUGAAAUAGCC---AGAGCAACCGACACGGCUCCCAAAA---AAACAAAACUUCA (((((((((((...(((.((...((((((------....)))))).)).)))....)))---).......)))))))..........---............. ( -22.41, z-score = -1.59, R) >droWil1.scaffold_181096 9800841 93 + 12416693 CGUGUUGGAGCAAAUUACUCGAGUGAAGUGUGUGUGUGCACUUCAAGAAUGAAAUAGCC---CAAGCAACUGACACGUCAAAGA------AAACAAAAACUC- (((((..(......(((.((...((((((((......)))))))).)).)))....((.---...))..)..))))).......------............- ( -22.70, z-score = -1.55, R) >droVir3.scaffold_12970 6081091 86 - 11907090 UGUGUUGAAGCAAAUUACACGAGUGAAGU------GCGCACUUCAAG-AUGAAAUAGUC----AAGCAACUGACACGCAAACAAC-----UCACUUUACAUC- .((((...........))))((((....(------(((...(((...-..)))...(((----(......)))).))))....))-----))..........- ( -17.40, z-score = -0.24, R) >droMoj3.scaffold_6473 1401353 86 + 16943266 UGUGUUGAGGCAAAUUACUCGAGUGAAGU------GCGCACUUCAAG-AUGAAAUAGUC----AAGCAACUGACACGCAAACAAC-----UCACUUUACAUC- ((((..(((........)))((((....(------(((...(((...-..)))...(((----(......)))).))))....))-----))....))))..- ( -19.10, z-score = -0.54, R) >consensus CGUGUCGUGGCAAAUUACUCGAGUGAAGU______GCGCACUUCAAGAAUGAAAUAGCC___AAAGCAACCGACACGGCUCCCAA_____AAACAAAACUUCA (((((((.......(((.((...((((((..........)))))).)).)))....((.......))...))))))).......................... (-13.03 = -12.62 + -0.41)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:28:31 2011