| Sequence ID | dm3.chrX |
|---|---|
| Location | 9,424,474 – 9,424,544 |
| Length | 70 |
| Max. P | 0.576159 |

| Location | 9,424,474 – 9,424,544 |
|---|---|
| Length | 70 |
| Sequences | 10 |
| Columns | 80 |
| Reading direction | forward |
| Mean pairwise identity | 79.52 |
| Shannon entropy | 0.36884 |
| G+C content | 0.43710 |
| Mean single sequence MFE | -14.99 |
| Consensus MFE | -7.34 |
| Energy contribution | -7.55 |
| Covariance contribution | 0.21 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.576159 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
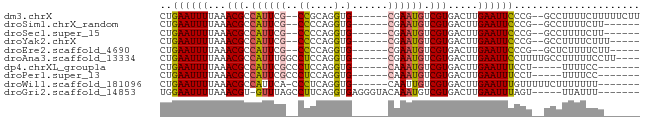
>dm3.chrX 9424474 70 + 22422827 CUGAAUUUUAAACGCCAUUCG--CCGCAGGUG------CGAAUGUCGUGACUUGAAUUCCCG--GCCUUUUCUUUUUCUU ..((((((...(((.((((((--((....).)------)))))).))).....))))))...--................ ( -16.50, z-score = -1.45, R) >droSim1.chrX_random 2708469 64 + 5698898 CUGAAUUUUAAACGCCAUUCG--CCCCAGGUG------CGAAUGUCGUGACUUGAAUUCCCG--GCCUUUUCUU------ ..((((((...(((.((((((--((....).)------)))))).))).....))))))...--..........------ ( -17.80, z-score = -2.78, R) >droSec1.super_15 188529 64 + 1954846 CUGAAUUUUAAACGCCAUUCG--CCCCAGGUG------CGAAUGUCGUGACUUGAAUUCCCG--GCCUUUUCUU------ ..((((((...(((.((((((--((....).)------)))))).))).....))))))...--..........------ ( -17.80, z-score = -2.78, R) >droYak2.chrX 18017186 65 + 21770863 CUGAAUUUUAAACGCCAUUCG--CCCCAGGUG------CGAAUGUCGUGACUUGAAUUCCCG--GCCUUUUCUUU----- ..((((((...(((.((((((--((....).)------)))))).))).....))))))...--...........----- ( -17.80, z-score = -2.79, R) >droEre2.scaffold_4690 13057301 65 - 18748788 CUGAAUUUUAAACGCCAUUCG--CCCCAGGUG------CGAAUGUCGUGACUUGAAUUCCCG--GCUCUUUUCUU----- ..((((((...(((.((((((--((....).)------)))))).))).....))))))...--...........----- ( -17.80, z-score = -2.74, R) >droAna3.scaffold_13334 1466966 70 - 1562580 CUGAAUUUUAAACGCCAUUUGGCCUCCAGGUG------CGAAUGUCGUGACUUGAAUUCCUUUUGCCUUUUUCCUU---- ..((((((...(((.(((((((((....))).------)))))).))).....)))))).................---- ( -15.10, z-score = -1.93, R) >dp4.chrXL_group1a 1597930 62 - 9151740 CUGAAUUUUAAACGCCAUUCGCCCUCCAGGUG------CAAAUGUCGUGACUUGAAUUUCCU-----UUUUCC------- ..(((.((((((((.((((.((((....)).)------).)))).)))...))))).)))..-----......------- ( -12.30, z-score = -2.17, R) >droPer1.super_13 94164 62 - 2293547 CUGAAUUUUAAACGCCAUUCGCCCUCCAGGUG------CAAAUGUCGUGACUUGAAUUUCCU-----UUUUCC------- ..(((.((((((((.((((.((((....)).)------).)))).)))...))))).)))..-----......------- ( -12.30, z-score = -2.17, R) >droWil1.scaffold_181096 9798427 66 - 12416693 CUGAAUUUUAAACGCCAUUCA-CCCUCAGGUG------CAAUUGUCGUGACUUGAAUUUGUUUUUCUUUUUUU------- ..((((((...((((.(((((-((....))))------.))).).))).....))))))..............------- ( -9.70, z-score = -0.87, R) >droGri2.scaffold_14853 929175 67 - 10151454 UGGAAUUUUAAACGU-GUUUAGCCUUCAGGUGAGGGUACAAAUGUCGUGACUUGAAUUUAGU-----UUAUUU------- ..((((((...((((-((((.((((((....))))))..))))).))).....))))))...-----......------- ( -12.80, z-score = -0.86, R) >consensus CUGAAUUUUAAACGCCAUUCG__CCCCAGGUG______CGAAUGUCGUGACUUGAAUUCCCG__GCCUUUUCUU______ ..((((((...(((.((((((.................)))))).))).....))))))..................... ( -7.34 = -7.55 + 0.21)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:28:30 2011