
| Sequence ID | dm3.chrX |
|---|---|
| Location | 9,398,390 – 9,398,453 |
| Length | 63 |
| Max. P | 0.999663 |

| Location | 9,398,390 – 9,398,453 |
|---|---|
| Length | 63 |
| Sequences | 11 |
| Columns | 64 |
| Reading direction | forward |
| Mean pairwise identity | 82.26 |
| Shannon entropy | 0.38587 |
| G+C content | 0.54134 |
| Mean single sequence MFE | -26.63 |
| Consensus MFE | -18.85 |
| Energy contribution | -17.93 |
| Covariance contribution | -0.92 |
| Combinations/Pair | 1.53 |
| Mean z-score | -2.99 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.17 |
| SVM RNA-class probability | 0.997732 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
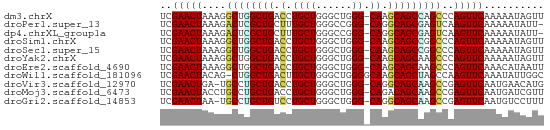
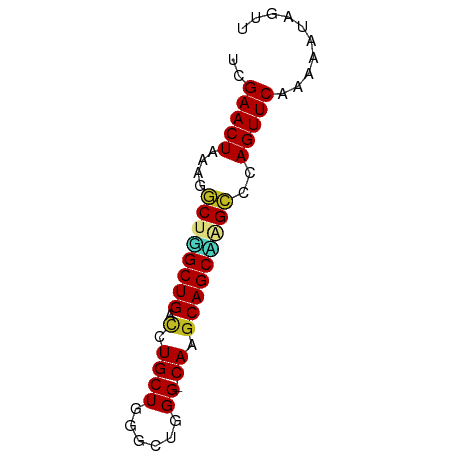
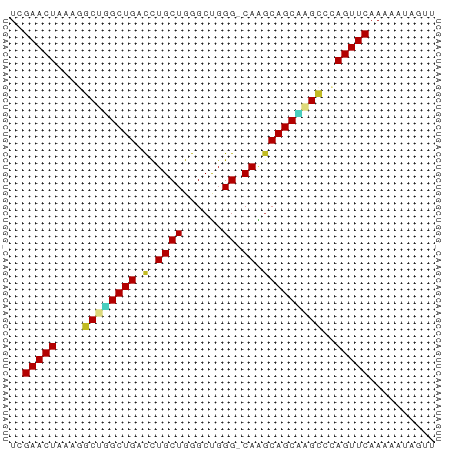
>dm3.chrX 9398390 63 + 22422827 UCGAACUAAAGGCUGGCUGACCUGCUGGGCUGGG-CAAGCAGCCAGCCCAGUUCAAAAAUAGUU ..(((((...(((((((((.(.((((......))-)).)))))))))).))))).......... ( -29.00, z-score = -3.25, R) >droPer1.super_13 61064 62 - 2293547 UCGAACUAAAGACUCGCUGCUUUGCUGGGCCGGG-CAGGCAGCGAGUCAAGUUCAAAAAUAUU- ..(((((...((((((((((((.(((......))-))))))))))))).))))).........- ( -30.10, z-score = -5.55, R) >dp4.chrXL_group1a 1564005 62 - 9151740 UCGAACUAAAGACUCGCUGCUUUGCUGGGCCGGG-CAGGCAGCGAGUCAAGUUCAAAAAUAUU- ..(((((...((((((((((((.(((......))-))))))))))))).))))).........- ( -30.10, z-score = -5.55, R) >droSim1.chrX 7493173 63 + 17042790 UCGAACUAAAGGCUGGCUGACCUGCUGGGCUGGG-CAAGCAGCCGGCCCAGUUCAAAAAUAGUU ..(((((...(((((((((.(.((((......))-)).)))))))))).))))).......... ( -28.30, z-score = -2.84, R) >droSec1.super_15 164838 63 + 1954846 UCGAACUAAAGGCUGGCUGACCUGCUGGGCUGGG-CAAGCAGCCGGCCCAGUUCAAAAAUAGUU ..(((((...(((((((((.(.((((......))-)).)))))))))).))))).......... ( -28.30, z-score = -2.84, R) >droYak2.chrX 17990407 63 + 21770863 UCGAACUAAAGGCUGGCUGACCUGCUGGGCUGGG-CAAGCAGCAAGCCCAGUUCAAAAAUAGUU ..(((((...((((.((((.(.((((......))-)).))))).)))).))))).......... ( -22.50, z-score = -1.32, R) >droEre2.scaffold_4690 13032668 63 - 18748788 UCGAACUAAAGGCUGGCUGACCUGCUGGGCUGGG-CAAGCAGCAAGCCCAGUUCAAACAUAAUU ..(((((...((((.((((.(.((((......))-)).))))).)))).))))).......... ( -22.50, z-score = -1.63, R) >droWil1.scaffold_181096 9764900 63 - 12416693 UCGAACUACAG-CUGGCUGACUUGCUGGGCUGGGGCAAGCAGCUAGCCAAGUUCAAAUAUUGGC ..(((((...(-(((((((.((((((.......))))))))))))))..))))).......... ( -27.40, z-score = -3.26, R) >droVir3.scaffold_12970 6035220 62 + 11907090 UCGAACUGA-UGCCUGCUGACCUGCUGGGCUGGG-CAGGCAGCAAGCCGAGUUCAAUGAACAUG ..(((((..-.((.(((((.((((((......))-))))))))).))..))))).......... ( -26.10, z-score = -2.77, R) >droMoj3.scaffold_6473 1353792 63 - 16943266 UCGAACUACCUGCCUGCUGACCUGCUGGGCUGGG-CAGACAGCAAGCCGAGUUCAAUGAUCGUU ..(((((....((.(((((..(((((......))-))).))))).))..))))).......... ( -21.40, z-score = -1.26, R) >droGri2.scaffold_14853 895212 62 - 10151454 UCGAACUAA-UGCCUGCUGUCCUGCUGGGCUGGG-CAGGCAGCAAGCCGAGUUCAAUGUCCUUU ..(((((..-.((.(((((.((((((......))-))))))))).))..))))).......... ( -27.20, z-score = -2.66, R) >consensus UCGAACUAAAGGCUGGCUGACCUGCUGGGCUGGG_CAAGCAGCAAGCCCAGUUCAAAAAUAGUU ..(((((....(((((((..((....))(((......))))))))))..))))).......... (-18.85 = -17.93 + -0.92)
| Location | 9,398,390 – 9,398,453 |
|---|---|
| Length | 63 |
| Sequences | 11 |
| Columns | 64 |
| Reading direction | reverse |
| Mean pairwise identity | 82.26 |
| Shannon entropy | 0.38587 |
| G+C content | 0.54134 |
| Mean single sequence MFE | -28.33 |
| Consensus MFE | -20.44 |
| Energy contribution | -20.31 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.35 |
| Mean z-score | -4.14 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.15 |
| SVM RNA-class probability | 0.999663 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
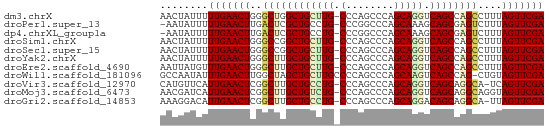
>dm3.chrX 9398390 63 - 22422827 AACUAUUUUUGAACUGGGCUGGCUGCUUG-CCCAGCCCAGCAGGUCAGCCAGCCUUUAGUUCGA ........(((((((((((((((((((((-(........))))).))))))))))..))))))) ( -33.30, z-score = -5.06, R) >droPer1.super_13 61064 62 + 2293547 -AAUAUUUUUGAACUUGACUCGCUGCCUG-CCCGGCCCAGCAAAGCAGCGAGUCUUUAGUUCGA -.......(((((((.((((((((((.((-(........)))..))))))))))...))))))) ( -28.30, z-score = -5.88, R) >dp4.chrXL_group1a 1564005 62 + 9151740 -AAUAUUUUUGAACUUGACUCGCUGCCUG-CCCGGCCCAGCAAAGCAGCGAGUCUUUAGUUCGA -.......(((((((.((((((((((.((-(........)))..))))))))))...))))))) ( -28.30, z-score = -5.88, R) >droSim1.chrX 7493173 63 - 17042790 AACUAUUUUUGAACUGGGCCGGCUGCUUG-CCCAGCCCAGCAGGUCAGCCAGCCUUUAGUUCGA ........(((((((((((.(((((((((-(........))))).))))).))))..))))))) ( -28.70, z-score = -3.72, R) >droSec1.super_15 164838 63 - 1954846 AACUAUUUUUGAACUGGGCCGGCUGCUUG-CCCAGCCCAGCAGGUCAGCCAGCCUUUAGUUCGA ........(((((((((((.(((((((((-(........))))).))))).))))..))))))) ( -28.70, z-score = -3.72, R) >droYak2.chrX 17990407 63 - 21770863 AACUAUUUUUGAACUGGGCUUGCUGCUUG-CCCAGCCCAGCAGGUCAGCCAGCCUUUAGUUCGA ........((((((((((((.((((((((-(........))))).)))).)))))..))))))) ( -26.80, z-score = -3.48, R) >droEre2.scaffold_4690 13032668 63 + 18748788 AAUUAUGUUUGAACUGGGCUUGCUGCUUG-CCCAGCCCAGCAGGUCAGCCAGCCUUUAGUUCGA ........((((((((((((.((((((((-(........))))).)))).)))))..))))))) ( -26.80, z-score = -3.32, R) >droWil1.scaffold_181096 9764900 63 + 12416693 GCCAAUAUUUGAACUUGGCUAGCUGCUUGCCCCAGCCCAGCAAGUCAGCCAG-CUGUAGUUCGA ........((((((((((((.(((((((((.........))))).)))).))-))).))))))) ( -24.00, z-score = -3.31, R) >droVir3.scaffold_12970 6035220 62 - 11907090 CAUGUUCAUUGAACUCGGCUUGCUGCCUG-CCCAGCCCAGCAGGUCAGCAGGCA-UCAGUUCGA ........(((((((..((((((((((((-(........))))).)))))))).-..))))))) ( -29.90, z-score = -4.17, R) >droMoj3.scaffold_6473 1353792 63 + 16943266 AACGAUCAUUGAACUCGGCUUGCUGUCUG-CCCAGCCCAGCAGGUCAGCAGGCAGGUAGUUCGA ........((((((((.((((((((((((-(........))))).))))))))..).))))))) ( -26.90, z-score = -2.85, R) >droGri2.scaffold_14853 895212 62 + 10151454 AAAGGACAUUGAACUCGGCUUGCUGCCUG-CCCAGCCCAGCAGGACAGCAGGCA-UUAGUUCGA ........(((((((..((((((((((((-(........))))).)))))))).-..))))))) ( -29.90, z-score = -4.17, R) >consensus AACUAUUUUUGAACUGGGCUUGCUGCUUG_CCCAGCCCAGCAGGUCAGCCAGCCUUUAGUUCGA ........(((((((..((((((((((((.(........))))).))))))))....))))))) (-20.44 = -20.31 + -0.13)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:28:29 2011