
| Sequence ID | dm3.chrX |
|---|---|
| Location | 9,390,142 – 9,390,254 |
| Length | 112 |
| Max. P | 0.737227 |

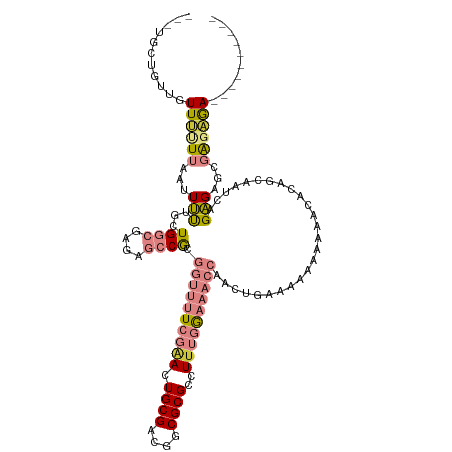
| Location | 9,390,142 – 9,390,254 |
|---|---|
| Length | 112 |
| Sequences | 7 |
| Columns | 122 |
| Reading direction | forward |
| Mean pairwise identity | 68.80 |
| Shannon entropy | 0.56575 |
| G+C content | 0.48630 |
| Mean single sequence MFE | -38.14 |
| Consensus MFE | -12.64 |
| Energy contribution | -15.40 |
| Covariance contribution | 2.76 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.732292 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 9390142 112 + 22422827 UGCUGUUGUUUUUUUUUUUAUUUUGCUGGCGAGAGCCGCGGUUUUCGAACUGCGACGGCGCGCCUUUGGAAACCAACUGAAAAAAAAAACACAGCAAUCAGAGAGCGAGAGA---------- ((((((.(((((((((((((....((.(((....)))))((((((((((.((((....))))..))))))))))...)))))))))))))))))))................---------- ( -42.30, z-score = -3.60, R) >droSim1.chrX 7484546 107 + 17042790 ---UGCUGUUGUUUUUUA--UUUUGCUGGCGAGAGCCGCGGUUUUCGAACUGCGACGGCGCGCCUUUGGAAACCAACUGAAAAAAAAAACACAGCAAUCAGAGAGCGAGAGA---------- ---((((((.(((((((.--((((((.(((....)))))((((((((((.((((....))))..))))))))))....)))).)))))))))))))................---------- ( -38.00, z-score = -2.66, R) >droSec1.super_15 156895 108 + 1954846 ---UGCUGUUGUUUUUUUAUUUU-GCUGGCGAGAGCCGCGGUUUUCGAACUGCGACGGCGCGCCUUUGGAAACCAACUGAAAAAAAAAACACAGCAAUCAGAGAGCGAGAGA---------- ---((((((.((((((((.((((-((.(((....)))))((((((((((.((((....))))..))))))))))....))))))))))))))))))................---------- ( -37.70, z-score = -2.50, R) >droYak2.chrX 17981671 108 + 21770863 ---UGUUGUUGUUUCUUAAUUUUUGUUGGCAAGAGCCGCGGUUUUCGAACUGCGACGGCGCGCCUUUGGAAACCAACUGAAAAAAAAA-CACAGAAAUCGGAGAGCGAGAGA---------- ---..((.(((((..(..((((((((.(((....)))..((((((((((.((((....))))..))))))))))..............-.))))))))..)..))))).)).---------- ( -31.30, z-score = -0.89, R) >droEre2.scaffold_4690 13024378 106 - 18748788 ---UGCUGUUGUUUUUUAAUUUU-GAUGGCGAGAGCCGCGGUUUUCGAACUGCGACGGCGCGCCUUUGGAAACCAACUGAAAAAAAA--CACAGAAAUCUGAGAGCGAGAGA---------- ---..((.((((((((..(((((-(.((((....)))))((((((((((.((((....))))..)))))))))).............--....)))))..)))))))).)).---------- ( -33.10, z-score = -2.07, R) >droVir3.scaffold_12970 6025048 111 + 11907090 ------UGUUGUUUUU--ACUGCUGCUGUUGCUGCUCAAAAGAGUGUGACUGCGACGGCGCGCCUAUGAAAAGCAACUGAAGC---AGCGACGCGACGUCGCGACUUGCCAACUCUGAUUGU ------.(((((((((--...(((((((((((.((((....))))......))))))))).)).....)))))))))..(((.---.((((((...))))))..)))............... ( -36.90, z-score = -0.98, R) >droMoj3.scaffold_6473 1344211 114 - 16943266 ------UGCUGUUGUU--GUUGCUGUUGUUGCUACUCAAUAGAGUGCGACUGCGACGGCGCGCCUUUGAAAAGCAACUGAGGCGGCAGCGACGCGGCGUCGCGUCUUGCCAACUCUGAUUGU ------.((.((((((--((((((...((((((..((((.((.(((((.(((...))))))))))))))..))))))...))))))))))))))((((........))))............ ( -47.70, z-score = -1.88, R) >consensus ___UGCUGUUGUUUUUUAAUUUUUGCUGGCGAGAGCCGCGGUUUUCGAACUGCGACGGCGCGCCUUUGGAAACCAACUGAAAAAAAAAACACAGCAAUCAGAGAGCGAGAGA__________ .....((((..((((((.........((((....)))).((((((((((.((((....))))..))))))))))........))))))..))))............................ (-12.64 = -15.40 + 2.76)

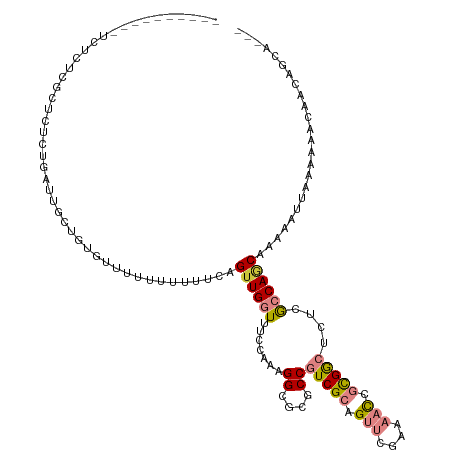
| Location | 9,390,142 – 9,390,254 |
|---|---|
| Length | 112 |
| Sequences | 7 |
| Columns | 122 |
| Reading direction | reverse |
| Mean pairwise identity | 68.80 |
| Shannon entropy | 0.56575 |
| G+C content | 0.48630 |
| Mean single sequence MFE | -31.89 |
| Consensus MFE | -12.00 |
| Energy contribution | -12.60 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.737227 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 9390142 112 - 22422827 ----------UCUCUCGCUCUCUGAUUGCUGUGUUUUUUUUUUCAGUUGGUUUCCAAAGGCGCGCCGUCGCAGUUCGAAAACCGCGGCUCUCGCCAGCAAAAUAAAAAAAAAAACAACAGCA ----------................(((((((((((((((((...((((...)))).((((.(..(((((.(((....))).))))).).)))).........))))))))))).)))))) ( -32.50, z-score = -2.37, R) >droSim1.chrX 7484546 107 - 17042790 ----------UCUCUCGCUCUCUGAUUGCUGUGUUUUUUUUUUCAGUUGGUUUCCAAAGGCGCGCCGUCGCAGUUCGAAAACCGCGGCUCUCGCCAGCAAAA--UAAAAAACAACAGCA--- ----------................(((((((((((((.(((..((((((.......((....))(((((.(((....))).)))))....)))))).)))--.))))))).))))))--- ( -30.90, z-score = -1.87, R) >droSec1.super_15 156895 108 - 1954846 ----------UCUCUCGCUCUCUGAUUGCUGUGUUUUUUUUUUCAGUUGGUUUCCAAAGGCGCGCCGUCGCAGUUCGAAAACCGCGGCUCUCGCCAGC-AAAAUAAAAAAACAACAGCA--- ----------................((((((((((((((.((..((((((.......((....))(((((.(((....))).)))))....))))))-..)).)))))))).))))))--- ( -32.20, z-score = -2.29, R) >droYak2.chrX 17981671 108 - 21770863 ----------UCUCUCGCUCUCCGAUUUCUGUG-UUUUUUUUUCAGUUGGUUUCCAAAGGCGCGCCGUCGCAGUUCGAAAACCGCGGCUCUUGCCAACAAAAAUUAAGAAACAACAACA--- ----------....(((.....)))....((((-(((((((((..((((((.......((....))(((((.(((....))).)))))....))))))..)))..))))))).)))...--- ( -23.10, z-score = -0.29, R) >droEre2.scaffold_4690 13024378 106 + 18748788 ----------UCUCUCGCUCUCAGAUUUCUGUG--UUUUUUUUCAGUUGGUUUCCAAAGGCGCGCCGUCGCAGUUCGAAAACCGCGGCUCUCGCCAUC-AAAAUUAAAAAACAACAGCA--- ----------......(((..(((....)))((--((((((.....(((((.......((((.(..(((((.(((....))).))))).).)))))))-))....))))))))..))).--- ( -25.81, z-score = -1.61, R) >droVir3.scaffold_12970 6025048 111 - 11907090 ACAAUCAGAGUUGGCAAGUCGCGACGUCGCGUCGCU---GCUUCAGUUGCUUUUCAUAGGCGCGCCGUCGCAGUCACACUCUUUUGAGCAGCAACAGCAGCAGU--AAAAACAACA------ .........((((....(.((((....)))).)(((---(((...((((((.......((((((....))).)))...(((....))).))))))))))))...--.....)))).------ ( -36.00, z-score = -1.47, R) >droMoj3.scaffold_6473 1344211 114 + 16943266 ACAAUCAGAGUUGGCAAGACGCGACGCCGCGUCGCUGCCGCCUCAGUUGCUUUUCAAAGGCGCGCCGUCGCAGUCGCACUCUAUUGAGUAGCAACAACAGCAAC--AACAACAGCA------ .......(((.(((((.((((((....))))))..))))).))).((((((.......((((((....))).)))((((((....)))).))......))))))--..........------ ( -42.70, z-score = -2.69, R) >consensus __________UCUCUCGCUCUCUGAUUGCUGUGUUUUUUUUUUCAGUUGGUUUCCAAAGGCGCGCCGUCGCAGUUCGAAAACCGCGGCUCUCGCCAGCAAAAAUUAAAAAACAACAGCA___ .............................................((((((.......((....))(((((.(((....))).)))))....))))))........................ (-12.00 = -12.60 + 0.60)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:28:26 2011