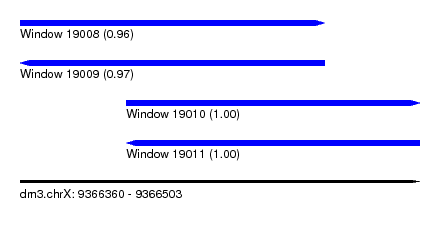
| Sequence ID | dm3.chrX |
|---|---|
| Location | 9,366,360 – 9,366,503 |
| Length | 143 |
| Max. P | 0.999522 |

| Location | 9,366,360 – 9,366,469 |
|---|---|
| Length | 109 |
| Sequences | 11 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 80.87 |
| Shannon entropy | 0.40389 |
| G+C content | 0.58823 |
| Mean single sequence MFE | -48.49 |
| Consensus MFE | -28.16 |
| Energy contribution | -27.66 |
| Covariance contribution | -0.49 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.957277 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
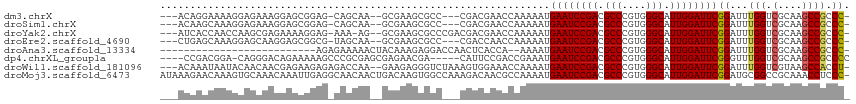
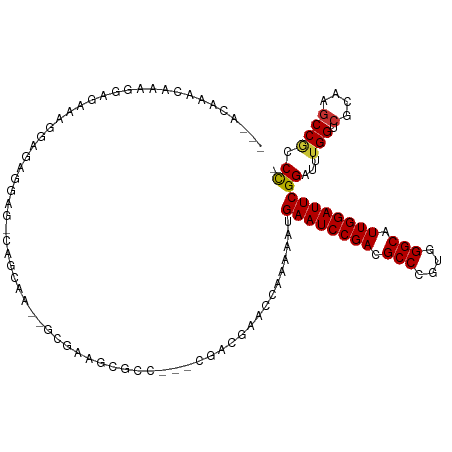
>dm3.chrX 9366360 109 + 22422827 GAUGGUGGUCGCCAGCGAGAACGCAUUCUGGCCUUGCUGGG---CGGCUUGCGACCAAAUCCGAAUCCAAUGCCCACGGGCGUCGGAUUCAUUUUUGGUUCGUCG---GGCGCUU-- (((....))).((((((((..((.....))..))))))))(---((.((.(((((((((...((((((.(((((....))))).))))))...))))).)))).)---).)))..-- ( -50.40, z-score = -2.79, R) >droSim1.chrX 7464596 109 + 17042790 GAUGGUGGUCGCCAGCGAGAACGCAUUCUGGCCUUGCUGGG---CGGCUUGCGACCAAAUCCGAAUCCAAUGCCCACGGGCGUCGGAUUCAUUUUUGGUUCGUCG---GGCGCUU-- (((....))).((((((((..((.....))..))))))))(---((.((.(((((((((...((((((.(((((....))))).))))))...))))).)))).)---).)))..-- ( -50.40, z-score = -2.79, R) >droSec1.super_15 133722 109 + 1954846 GAUGGUGGUCGCCAGCGAGAACGCAUUCUGGCCUUGCUGGG---CGGCUUGCGACCAAAUCCGAAUCCAAUGCCCACGGGCGUCGGAUUCAUUUUUGGUUCGUCG---GGCGCUU-- (((....))).((((((((..((.....))..))))))))(---((.((.(((((((((...((((((.(((((....))))).))))))...))))).)))).)---).)))..-- ( -50.40, z-score = -2.79, R) >droYak2.chrX 17955835 112 + 21770863 GACGGUGGCCGCCAGCGAGAACGCAUUCUGGCCUUGCUGGG---CGGCUUGCGACCAAAUCCGAAUCCAAUGCCCACGGGCGUCGGAUUCAUUUUUGGUUCGUCGUCGGGCGCUU-- ((((..(((((((((((((..((.....))..)))))).))---))))).(((((((((...((((((.(((((....))))).))))))...))))).))))))))........-- ( -55.10, z-score = -3.32, R) >droEre2.scaffold_4690 13001129 109 - 18748788 GUUGGUGGUCGCCAGCGAGAACGCAUUCUGGCCUUGCUGGG---CGGCUUGCGACCAAAUCCGAAUCCAAUGCCCACGGGCGUCGGAUUCAUUUUUGGUUGGUCG---GGCGCUU-- ...((((.((((((((.((((((((..(((.((.....)).---)))..)))).........((((((.(((((....))))).))))))..)))).))))).))---).)))).-- ( -50.00, z-score = -2.56, R) >droAna3.scaffold_13334 1397998 103 - 1562580 GAUAAUGGCCGCCAACGGGAACGCAUUCUGGCCUUGUUGGG---CGGCUUGCGACCAAAUCCGAAUCCAAUGCCCACGGGCGUCGGAUUCAUUUUUGGUGAGUUGG----------- ......(((((((((((((..((.....))..)))))).))---))))).((.((((((...((((((.(((((....))))).))))))...))))))..))...----------- ( -43.80, z-score = -2.65, R) >dp4.chrXL_group1a 1524834 112 - 9151740 GGUCGGGGUCGCCAGCGGGAGCGCAUUUUUGCGUUGUUGGGGGGCGGCUUGCGACCAAACCCGAAUCCAAUGCCCACGGGCGUCGGAUUCAUUUCGGUCGGAAUGUCGUUCU----- (((((((((((((..(((.((((((....)))))).)))...)))))))).)))))......((((((.(((((....))))).))))))(((((....)))))........----- ( -52.50, z-score = -2.47, R) >droWil1.scaffold_181096 9728736 112 - 12416693 ACUGGUGGCGGCCAACGUGAGCGCAUUUUGGCCUUAUUAGG---UGGCUUACGACCAAAUCCGAAUCCAAUGCCCACGGGCGUCGGAUUCAUUUUGGUUUCCACUUUAGACCCUC-- .(((((((.((((((.(((....))).)))))))))))))(---(((.....(((((((...((((((.(((((....))))).))))))..))))))).))))...........-- ( -45.40, z-score = -3.88, R) >droVir3.scaffold_12970 5997141 114 + 11907090 GUUGGUGGCGGCCAUCGGGAACGCAUCUUGGCAUUGUUGGG---UGGUUUGCGGCCGCAUCCGAAUCCAAUGCCCACGGGCCUCGGAUUCAUUUUGGCGUUGUCUUUGGCCACUUGU ......((.(((((..((.(((((..((..((...))..))---.((..(((....))).))((((((...(((....)))...))))))......))))).))..))))).))... ( -42.40, z-score = -0.05, R) >droMoj3.scaffold_6473 1315704 114 - 16943266 GUUGGUGGGGGCCACCGGGAGCGCUUCUUGGCAUUGUUGGG---AGGUUUGCGGCCGCAUCCGAAUCCAAUGCCCACGGGCGUCGGAUUCAUUUUGGCGUUGUCUUUGGCCACUUGU ....(..(((((((..((.(((((((((..((...))..))---)((..(((....))).))((((((.(((((....))))).)))))).....)))))).))..))))).))..) ( -49.60, z-score = -1.67, R) >droGri2.scaffold_14853 859132 114 - 10151454 GGUGGUGGCGGCCAUCGGGAACGCAUCUUGGCAUUGUUGGG---UGGUUUGCGGCCGCAUCCGAAUCCAAUGCCCACGGGCCUCGGAUUCAUUUUGGCGUUGUCUUUGGCCACUUGU ((((((.((((((...(.((((.((.((..((...))..))---)))))).)))))))....((((((...(((....)))...))))))..................))))))... ( -43.40, z-score = -0.03, R) >consensus GAUGGUGGCCGCCAGCGAGAACGCAUUCUGGCCUUGUUGGG___CGGCUUGCGACCAAAUCCGAAUCCAAUGCCCACGGGCGUCGGAUUCAUUUUUGGUUCGUCGU__GGCGCUU__ ..((((.((.(((..((....))...((..((...))..))....)))..)).)))).....((((((.(((((....))))).))))))........................... (-28.16 = -27.66 + -0.49)
| Location | 9,366,360 – 9,366,469 |
|---|---|
| Length | 109 |
| Sequences | 11 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 80.87 |
| Shannon entropy | 0.40389 |
| G+C content | 0.58823 |
| Mean single sequence MFE | -42.13 |
| Consensus MFE | -25.93 |
| Energy contribution | -26.19 |
| Covariance contribution | 0.26 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.88 |
| SVM RNA-class probability | 0.973166 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 9366360 109 - 22422827 --AAGCGCC---CGACGAACCAAAAAUGAAUCCGACGCCCGUGGGCAUUGGAUUCGGAUUUGGUCGCAAGCCG---CCCAGCAAGGCCAGAAUGCGUUCUCGCUGGCGACCACCAUC --...((((---.(.(((((((((..(((((((((.(((....))).)))))))))..))))))((((..(.(---((......)))..)..))))...)))).))))......... ( -43.10, z-score = -2.35, R) >droSim1.chrX 7464596 109 - 17042790 --AAGCGCC---CGACGAACCAAAAAUGAAUCCGACGCCCGUGGGCAUUGGAUUCGGAUUUGGUCGCAAGCCG---CCCAGCAAGGCCAGAAUGCGUUCUCGCUGGCGACCACCAUC --...((((---.(.(((((((((..(((((((((.(((....))).)))))))))..))))))((((..(.(---((......)))..)..))))...)))).))))......... ( -43.10, z-score = -2.35, R) >droSec1.super_15 133722 109 - 1954846 --AAGCGCC---CGACGAACCAAAAAUGAAUCCGACGCCCGUGGGCAUUGGAUUCGGAUUUGGUCGCAAGCCG---CCCAGCAAGGCCAGAAUGCGUUCUCGCUGGCGACCACCAUC --...((((---.(.(((((((((..(((((((((.(((....))).)))))))))..))))))((((..(.(---((......)))..)..))))...)))).))))......... ( -43.10, z-score = -2.35, R) >droYak2.chrX 17955835 112 - 21770863 --AAGCGCCCGACGACGAACCAAAAAUGAAUCCGACGCCCGUGGGCAUUGGAUUCGGAUUUGGUCGCAAGCCG---CCCAGCAAGGCCAGAAUGCGUUCUCGCUGGCGGCCACCGUC --........((((.(((.(((((..(((((((((.(((....))).)))))))))..))))))))...((((---((.(((.((..(.......)..)).)))))))))...)))) ( -49.70, z-score = -2.94, R) >droEre2.scaffold_4690 13001129 109 + 18748788 --AAGCGCC---CGACCAACCAAAAAUGAAUCCGACGCCCGUGGGCAUUGGAUUCGGAUUUGGUCGCAAGCCG---CCCAGCAAGGCCAGAAUGCGUUCUCGCUGGCGACCACCAAC --..((...---(((((((.......(((((((((.(((....))).)))))))))...)))))))...))((---((.(((.((..(.......)..)).)))))))......... ( -41.50, z-score = -2.22, R) >droAna3.scaffold_13334 1397998 103 + 1562580 -----------CCAACUCACCAAAAAUGAAUCCGACGCCCGUGGGCAUUGGAUUCGGAUUUGGUCGCAAGCCG---CCCAACAAGGCCAGAAUGCGUUCCCGUUGGCGGCCAUUAUC -----------.......((((((..(((((((((.(((....))).)))))))))..)))))).....((((---((.(((((.((......)).))...)))))))))....... ( -40.10, z-score = -2.72, R) >dp4.chrXL_group1a 1524834 112 + 9151740 -----AGAACGACAUUCCGACCGAAAUGAAUCCGACGCCCGUGGGCAUUGGAUUCGGGUUUGGUCGCAAGCCGCCCCCCAACAACGCAAAAAUGCGCUCCCGCUGGCGACCCCGACC -----....((......((((((((.(((((((((.(((....))).)))))))))..)))))))).....((((..........(((....)))((....)).))))....))... ( -40.20, z-score = -1.83, R) >droWil1.scaffold_181096 9728736 112 + 12416693 --GAGGGUCUAAAGUGGAAACCAAAAUGAAUCCGACGCCCGUGGGCAUUGGAUUCGGAUUUGGUCGUAAGCCA---CCUAAUAAGGCCAAAAUGCGCUCACGUUGGCCGCCACCAGU --(..(((.....((((..((((((.(((((((((.(((....))).)))))))))..))))))......)))---).......((((((..((....))..)))))))))..)... ( -43.10, z-score = -2.91, R) >droVir3.scaffold_12970 5997141 114 - 11907090 ACAAGUGGCCAAAGACAACGCCAAAAUGAAUCCGAGGCCCGUGGGCAUUGGAUUCGGAUGCGGCCGCAAACCA---CCCAACAAUGCCAAGAUGCGUUCCCGAUGGCCGCCACCAAC ....(((((((..(..(((((......((((((((.(((....))).))))))))((.(((....)))..)).---.................)))))..)..)))))))....... ( -40.70, z-score = -2.04, R) >droMoj3.scaffold_6473 1315704 114 + 16943266 ACAAGUGGCCAAAGACAACGCCAAAAUGAAUCCGACGCCCGUGGGCAUUGGAUUCGGAUGCGGCCGCAAACCU---CCCAACAAUGCCAAGAAGCGCUCCCGGUGGCCCCCACCAAC ....((((((...(....)((.....(((((((((.(((....))).)))))))))...))))))))......---.........((......))......(((((...)))))... ( -38.10, z-score = -1.37, R) >droGri2.scaffold_14853 859132 114 + 10151454 ACAAGUGGCCAAAGACAACGCCAAAAUGAAUCCGAGGCCCGUGGGCAUUGGAUUCGGAUGCGGCCGCAAACCA---CCCAACAAUGCCAAGAUGCGUUCCCGAUGGCCGCCACCACC ....(((((((..(..(((((......((((((((.(((....))).))))))))((.(((....)))..)).---.................)))))..)..)))))))....... ( -40.70, z-score = -1.93, R) >consensus __AAGCGCC__ACGACGAACCAAAAAUGAAUCCGACGCCCGUGGGCAUUGGAUUCGGAUUUGGUCGCAAGCCG___CCCAACAAGGCCAGAAUGCGUUCCCGCUGGCGACCACCAUC ...........................((((((((.(((....))).))))))))((....((.......)).............((((....(((....)))))))..))...... (-25.93 = -26.19 + 0.26)

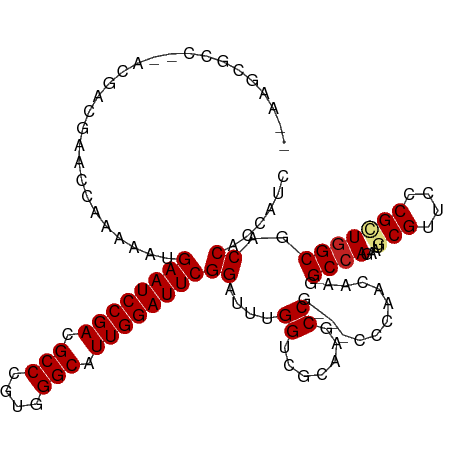
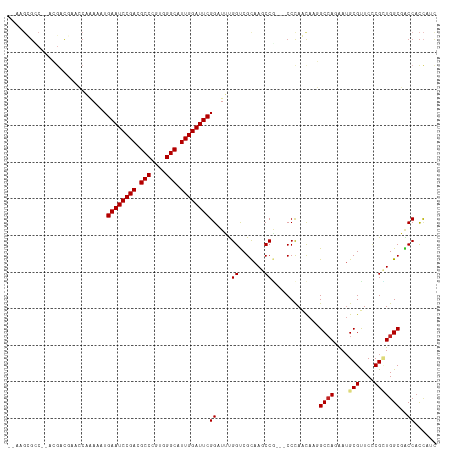
| Location | 9,366,398 – 9,366,503 |
|---|---|
| Length | 105 |
| Sequences | 8 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 68.80 |
| Shannon entropy | 0.65929 |
| G+C content | 0.55151 |
| Mean single sequence MFE | -39.88 |
| Consensus MFE | -19.76 |
| Energy contribution | -19.79 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.97 |
| SVM RNA-class probability | 0.999522 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chrX 9366398 105 + 22422827 -GGGCGGCUUGCGACCAAAUCCGAAUCCAAUGCCCACGGGCGUCGGAUUCAUUUUUGGUUCGUCG---GGCGCUUCGC--UUGCUG-CUCCGCUCCUUUCUCCUUUUCCUGU--- -((((((...(((((((((...((((((.(((((....))))).))))))...))))))).(.((---((((...)))--)))).)-).)))))).................--- ( -41.60, z-score = -3.06, R) >droSim1.chrX 7464634 105 + 17042790 -GGGCGGCUUGCGACCAAAUCCGAAUCCAAUGCCCACGGGCGUCGGAUUCAUUUUUGGUUCGUCG---GGCGCUUCGC--UUGCUG-CUCCGCUCCUUUCUCCUUUGCUUGU--- -((((((...(((((((((...((((((.(((((....))))).))))))...))))))).(.((---((((...)))--)))).)-).)))))).................--- ( -41.60, z-score = -2.76, R) >droYak2.chrX 17955873 107 + 21770863 -GGGCGGCUUGCGACCAAAUCCGAAUCCAAUGCCCACGGGCGUCGGAUUCAUUUUUGGUUCGUCGUCGGGCGCUUCGC--CU-UUU-CUCCUUUUCUCGCUUGGUUGGUGAU--- -.((((((....(((((((...((((((.(((((....))))).))))))...))))))).))))))(((((...)))--))-...-.........(((((.....))))).--- ( -41.80, z-score = -2.36, R) >droEre2.scaffold_4690 13001167 105 - 18748788 -GGGCGGCUUGCGACCAAAUCCGAAUCCAAUGCCCACGGGCGUCGGAUUCAUUUUUGGUUGGUCG---GGCGCUUCGC--UUGCUA-CGCCGCUCCUUGCUCCUUUGCUCAG--- -(((((((...((((((((...((((((.(((((....))))).))))))...))))))))(.((---((((...)))--))))..-.)))))))...((......))....--- ( -47.30, z-score = -3.80, R) >droAna3.scaffold_13334 1398036 86 - 1562580 -GGGCGGCUUGCGACCAAAUCCGAAUCCAAUGCCCACGGGCGUCGGAUUCAUUUU--UGGUGAGUUGGUCCUCUUUGUAGUUUUUCUCU-------------------------- -(((.((((.((.((((((...((((((.(((((....))))).))))))...))--))))..)).)))))))................-------------------------- ( -32.80, z-score = -3.05, R) >dp4.chrXL_group1a 1524874 105 - 9151740 GGGGCGGCUUGCGACCAAACCCGAAUCCAAUGCCCACGGGCGUCGGAUUCAUUUCGGUCGGAAUG-----UCGUUCUCGCUCGCGGGCUUUUUCUGUCCCUG-UCCGUCGG---- ((((((((((.(((((......((((((.(((((....))))).)))))).....))))).)).)-----)))))))....(((((((.............)-)))).)).---- ( -44.12, z-score = -2.02, R) >droWil1.scaffold_181096 9728774 109 - 12416693 -AGGUGGCUUACGACCAAAUCCGAAUCCAAUGCCCACGGGCGUCGGAUUCAUUUUGGUUUCCACUUUAGACCCUCUUC--UUGGUCUCUCUUCUCGUUGUUGUAUUAUUUGU--- -((((((.....(((((((...((((((.(((((....))))).))))))..))))))).)))))).(((((......--..))))).........................--- ( -34.40, z-score = -3.66, R) >droMoj3.scaffold_6473 1315742 114 - 16943266 -GGGAGGUUUGCGGCCGCAUCCGAAUCCAAUGCCCACGGGCGUCGGAUUCAUUUUGGCGUUGUCUUUGGCCACUUGUCAGUUGUUGCCUCAAUUUGUUUGCACUUUGUUCUUUAU -..(((((..(((((.(((...((((((.(((((....))))).))))))....((((..........))))..)))..))))).)))))......................... ( -35.40, z-score = -1.08, R) >consensus _GGGCGGCUUGCGACCAAAUCCGAAUCCAAUGCCCACGGGCGUCGGAUUCAUUUUUGGUUCGUCG___GGCGCUUCGC__UUGCUG_CUCCGCUCCUUGCUCCUUUGCUUGU___ ..((((.....)).))......((((((.(((((....))))).))))))................................................................. (-19.76 = -19.79 + 0.03)

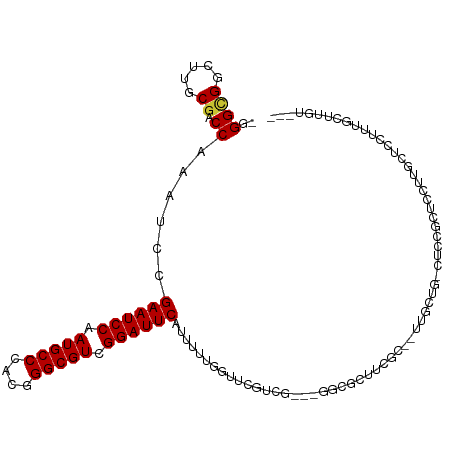
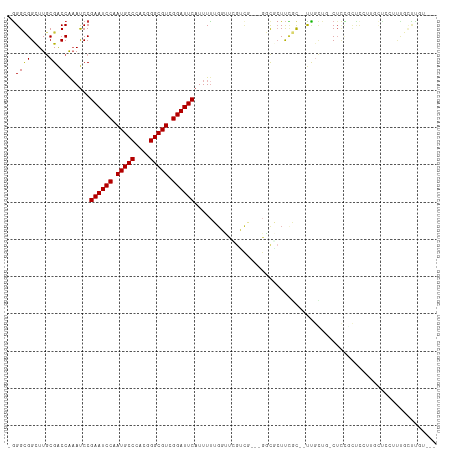
| Location | 9,366,398 – 9,366,503 |
|---|---|
| Length | 105 |
| Sequences | 8 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 68.80 |
| Shannon entropy | 0.65929 |
| G+C content | 0.55151 |
| Mean single sequence MFE | -36.66 |
| Consensus MFE | -20.61 |
| Energy contribution | -20.76 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.95 |
| SVM RNA-class probability | 0.999504 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chrX 9366398 105 - 22422827 ---ACAGGAAAAGGAGAAAGGAGCGGAG-CAGCAA--GCGAAGCGCC---CGACGAACCAAAAAUGAAUCCGACGCCCGUGGGCAUUGGAUUCGGAUUUGGUCGCAAGCCGCCC- ---...((....((.....((.(((..(-(.....--))....))))---)(.(((.(((((..(((((((((.(((....))).)))))))))..)))))))))...)).)).- ( -38.70, z-score = -2.27, R) >droSim1.chrX 7464634 105 - 17042790 ---ACAAGCAAAGGAGAAAGGAGCGGAG-CAGCAA--GCGAAGCGCC---CGACGAACCAAAAAUGAAUCCGACGCCCGUGGGCAUUGGAUUCGGAUUUGGUCGCAAGCCGCCC- ---................((.((((.(-(.((..--.....)))).---.(.(((.(((((..(((((((((.(((....))).)))))))))..)))))))))...))))))- ( -38.40, z-score = -2.25, R) >droYak2.chrX 17955873 107 - 21770863 ---AUCACCAACCAAGCGAGAAAAGGAG-AAA-AG--GCGAAGCGCCCGACGACGAACCAAAAAUGAAUCCGACGCCCGUGGGCAUUGGAUUCGGAUUUGGUCGCAAGCCGCCC- ---............(((..........-...-.(--(((...))))....(.(((.(((((..(((((((((.(((....))).)))))))))..)))))))))....)))..- ( -35.60, z-score = -2.06, R) >droEre2.scaffold_4690 13001167 105 + 18748788 ---CUGAGCAAAGGAGCAAGGAGCGGCG-UAGCAA--GCGAAGCGCC---CGACCAACCAAAAAUGAAUCCGACGCCCGUGGGCAUUGGAUUCGGAUUUGGUCGCAAGCCGCCC- ---....((......))..((.(((((.-..((..--(((...))).---.((((((.......(((((((((.(((....))).)))))))))...))))))))..)))))))- ( -42.80, z-score = -2.71, R) >droAna3.scaffold_13334 1398036 86 + 1562580 --------------------------AGAGAAAAACUACAAAGAGGACCAACUCACCA--AAAAUGAAUCCGACGCCCGUGGGCAUUGGAUUCGGAUUUGGUCGCAAGCCGCCC- --------------------------................(((......)))((((--((..(((((((((.(((....))).)))))))))..)))))).((.....))..- ( -28.70, z-score = -2.95, R) >dp4.chrXL_group1a 1524874 105 + 9151740 ----CCGACGGA-CAGGGACAGAAAAAGCCCGCGAGCGAGAACGA-----CAUUCCGACCGAAAUGAAUCCGACGCCCGUGGGCAUUGGAUUCGGGUUUGGUCGCAAGCCGCCCC ----.....((.-..(((.(.......))))(((.((..(((...-----..)))((((((((.(((((((((.(((....))).)))))))))..))))))))...))))))). ( -40.60, z-score = -1.85, R) >droWil1.scaffold_181096 9728774 109 + 12416693 ---ACAAAUAAUACAACAACGAGAAGAGAGACCAA--GAAGAGGGUCUAAAGUGGAAACCAAAAUGAAUCCGACGCCCGUGGGCAUUGGAUUCGGAUUUGGUCGUAAGCCACCU- ---.........................(((((..--......)))))...((((..((((((.(((((((((.(((....))).)))))))))..))))))......))))..- ( -35.10, z-score = -3.78, R) >droMoj3.scaffold_6473 1315742 114 + 16943266 AUAAAGAACAAAGUGCAAACAAAUUGAGGCAACAACUGACAAGUGGCCAAAGACAACGCCAAAAUGAAUCCGACGCCCGUGGGCAUUGGAUUCGGAUGCGGCCGCAAACCUCCC- .........................((((((.....))....((((((...(....)((.....(((((((((.(((....))).)))))))))...))))))))...))))..- ( -33.40, z-score = -2.00, R) >consensus ___ACAAACAAAGGAGAAAGGAGAGGAG_CAGCAA__GCGAAGCGCC___CGACGAACCAAAAAUGAAUCCGACGCCCGUGGGCAUUGGAUUCGGAUUUGGUCGCAAGCCGCCC_ .................................................................((((((((.(((....))).))))))))((...(((.(....)))))).. (-20.61 = -20.76 + 0.16)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:28:22 2011