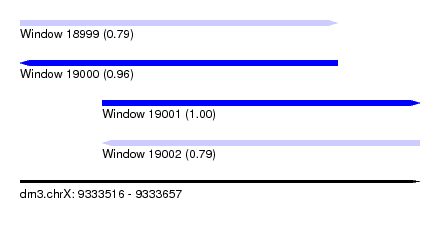
| Sequence ID | dm3.chrX |
|---|---|
| Location | 9,333,516 – 9,333,657 |
| Length | 141 |
| Max. P | 0.997893 |

| Location | 9,333,516 – 9,333,628 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 83.36 |
| Shannon entropy | 0.33076 |
| G+C content | 0.49762 |
| Mean single sequence MFE | -22.13 |
| Consensus MFE | -15.96 |
| Energy contribution | -16.35 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.790527 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
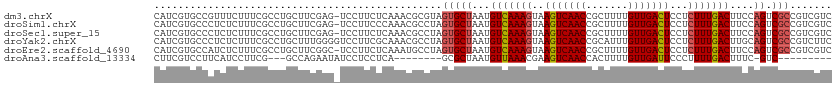
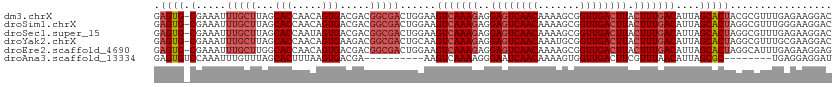
>dm3.chrX 9333516 112 + 22422827 CAUCGUGCCGUUUCUUUCGCCUGCUUCGAG-UCCUUCUCAAACGCGUAGUGCUAAUGUCAAAGUAAGUCAACCGCUUUUGUUGACUCCUCUUUGACUUCCAGUCGCCGUCGUC ...((.((..........((((((((.(((-.....))).))...)))).))....(((((((..(((((((.......)))))))...)))))))........))))..... ( -25.30, z-score = -1.99, R) >droSim1.chrX 7430585 112 + 17042790 CAUCGUGCCCUCUCUUUCGCCUGCUUCGAG-UCCUUCCCAAACGCCUAGUGCUAAUGUCAAAGUAAGUCAACCGCUUUUGUUGACUCCUCUUUGACUUCCAGUCGCCGUCGUC ..(((.((...(......)...))..))).-..........(((....(((((...(((((((..(((((((.......)))))))...)))))))....)).)))...))). ( -21.40, z-score = -1.72, R) >droSec1.super_15 102195 112 + 1954846 CAUCGUGCCCUCUCUUUCGCCUGCUUCGAG-UCCUUCUCAAACGCCUAGUGCUAAUGUCAAAGUAAGUCAACCGCUUUUGUUGACUCCUCUUUGACUUCCAGUCGCCGUCGUC ...((.((..........((((((((.(((-.....))).)).))..)).))....(((((((..(((((((.......)))))))...)))))))........))))..... ( -23.90, z-score = -2.42, R) >droYak2.chrX 17923132 113 + 21770863 CAUCGUGCCCUCUCUUUCGCCUGCUUUGGGGUCCUUCGCAAACGCCUAGUGCUAAUGUCAAAGUAAGUCAACCGCAUUUGUUGACUCCUCUUUGACUUGCAGUCGCCGUCUUC .................((.((((.....((..((..((....))..))..))...(((((((..(((((((.......)))))))...)))))))..)))).))........ ( -27.60, z-score = -1.56, R) >droEre2.scaffold_4690 12967393 112 - 18748788 CAUCGUGCCAUCUCUUUCGCCUGCUUCGGC-UCCUUCUCAAAUGCCUAGUGCUAAUGUCAAAGUAAGUCAACCGCUUUUGUUGACUCCUCUUUGACUUCCAGUCGCCGUCGUC ...((.((..........((..(((..(((-............))).)))))....(((((((..(((((((.......)))))))...)))))))........))))..... ( -24.50, z-score = -2.63, R) >droAna3.scaffold_13334 1361210 92 - 1562580 CUUCGUCCUUCAUCCUUCG---GCCAGAAUAUCCUCCUCA--------GCGCUAAUGUUAAACGAAGUCAACCACUUUUGUUGAUUCCCUUUUGACUUUC-GUC--------- ........(((...(....---)...)))...........--------(((.....((((((.(.(((((((.......)))))))..).))))))...)-)).--------- ( -10.10, z-score = 0.50, R) >consensus CAUCGUGCCCUCUCUUUCGCCUGCUUCGAG_UCCUUCUCAAACGCCUAGUGCUAAUGUCAAAGUAAGUCAACCGCUUUUGUUGACUCCUCUUUGACUUCCAGUCGCCGUCGUC ................................................(((((...(((((((..(((((((.......)))))))...)))))))....)).)))....... (-15.96 = -16.35 + 0.39)
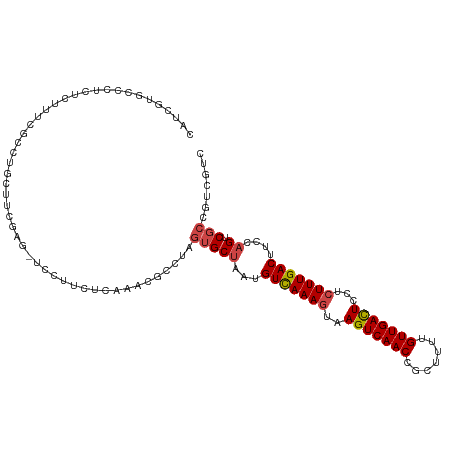
| Location | 9,333,516 – 9,333,628 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 83.36 |
| Shannon entropy | 0.33076 |
| G+C content | 0.49762 |
| Mean single sequence MFE | -31.68 |
| Consensus MFE | -19.51 |
| Energy contribution | -21.35 |
| Covariance contribution | 1.84 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.964744 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 9333516 112 - 22422827 GACGACGGCGACUGGAAGUCAAAGAGGAGUCAACAAAAGCGGUUGACUUACUUUGACAUUAGCACUACGCGUUUGAGAAGGA-CUCGAAGCAGGCGAAAGAAACGGCACGAUG .....((((........(((((((..((((((((.......)))))))).)))))))....((.((..((.((((((.....-))))))))))))..........)).))... ( -31.70, z-score = -2.08, R) >droSim1.chrX 7430585 112 - 17042790 GACGACGGCGACUGGAAGUCAAAGAGGAGUCAACAAAAGCGGUUGACUUACUUUGACAUUAGCACUAGGCGUUUGGGAAGGA-CUCGAAGCAGGCGAAAGAGAGGGCACGAUG .....((((..((....(((((((..((((((((.......)))))))).)))))))....((.....((.((((((.....-))))))))..)).......)).)).))... ( -31.20, z-score = -1.92, R) >droSec1.super_15 102195 112 - 1954846 GACGACGGCGACUGGAAGUCAAAGAGGAGUCAACAAAAGCGGUUGACUUACUUUGACAUUAGCACUAGGCGUUUGAGAAGGA-CUCGAAGCAGGCGAAAGAGAGGGCACGAUG .....((((..((....(((((((..((((((((.......)))))))).)))))))....((.....((.((((((.....-))))))))..)).......)).)).))... ( -32.10, z-score = -2.26, R) >droYak2.chrX 17923132 113 - 21770863 GAAGACGGCGACUGCAAGUCAAAGAGGAGUCAACAAAUGCGGUUGACUUACUUUGACAUUAGCACUAGGCGUUUGCGAAGGACCCCAAAGCAGGCGAAAGAGAGGGCACGAUG .....((((...((((((((((((..((((((((.......)))))))).))))))).)).)))((...(((((((...((...))...)))))))....))...)).))... ( -34.70, z-score = -2.39, R) >droEre2.scaffold_4690 12967393 112 + 18748788 GACGACGGCGACUGGAAGUCAAAGAGGAGUCAACAAAAGCGGUUGACUUACUUUGACAUUAGCACUAGGCAUUUGAGAAGGA-GCCGAAGCAGGCGAAAGAGAUGGCACGAUG ..((.((((..((....(((((((..((((((((.......)))))))).)))))))....((.....))........))..-))))..((...(....).....)).))... ( -35.10, z-score = -3.89, R) >droAna3.scaffold_13334 1361210 92 + 1562580 ---------GAC-GAAAGUCAAAAGGGAAUCAACAAAAGUGGUUGACUUCGUUUAACAUUAGCGC--------UGAGGAGGAUAUUCUGGC---CGAAGGAUGAAGGACGAAG ---------(((-....))).........(((((.......)))))((((((((..((((..((.--------(.((((.....)))).).---))...))))..)))))))) ( -25.30, z-score = -2.82, R) >consensus GACGACGGCGACUGGAAGUCAAAGAGGAGUCAACAAAAGCGGUUGACUUACUUUGACAUUAGCACUAGGCGUUUGAGAAGGA_CUCGAAGCAGGCGAAAGAGAGGGCACGAUG .................(((((((..((((((((.......)))))))).)))))))....((.....((.((((((......))))))))...(....).....))...... (-19.51 = -21.35 + 1.84)

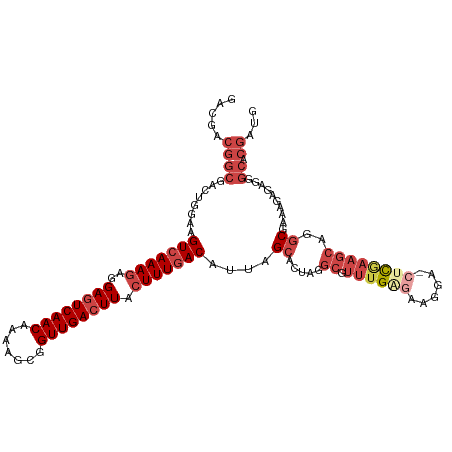
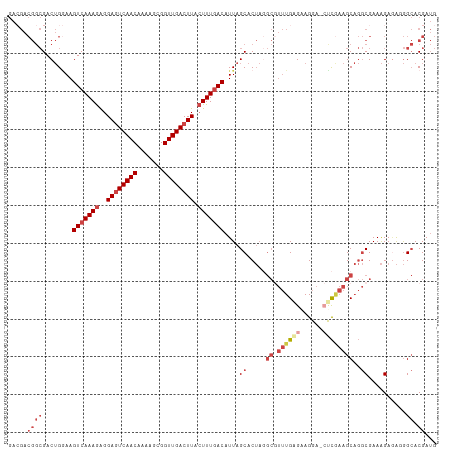
| Location | 9,333,545 – 9,333,657 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 87.00 |
| Shannon entropy | 0.25629 |
| G+C content | 0.45926 |
| Mean single sequence MFE | -28.46 |
| Consensus MFE | -23.02 |
| Energy contribution | -25.13 |
| Covariance contribution | 2.11 |
| Combinations/Pair | 1.13 |
| Mean z-score | -3.44 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.20 |
| SVM RNA-class probability | 0.997893 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
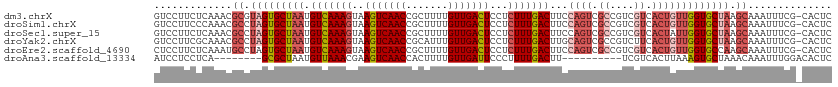
>dm3.chrX 9333545 112 + 22422827 GUCCUUCUCAAACGCGUAGUGCUAAUGUCAAAGUAAGUCAACCGCUUUUGUUGACUCCUCUUUGACUUCCAGUCGCCGUCGUCACUGUUGGUGCUAAGCAAAUUUCG-CACUC .............((.(((..((((.(((((((..(((((((.......)))))))...)))))))...((((.((....)).))))))))..))).))........-..... ( -32.00, z-score = -3.92, R) >droSim1.chrX 7430614 112 + 17042790 GUCCUUCCCAAACGCCUAGUGCUAAUGUCAAAGUAAGUCAACCGCUUUUGUUGACUCCUCUUUGACUUCCAGUCGCCGUCGUCACUGUUGGUGCUAAGCAAAUUUCG-CACUC .............((.(((..((((.(((((((..(((((((.......)))))))...)))))))...((((.((....)).))))))))..))).))........-..... ( -32.00, z-score = -4.52, R) >droSec1.super_15 102224 112 + 1954846 GUCCUUCUCAAACGCCUAGUGCUAAUGUCAAAGUAAGUCAACCGCUUUUGUUGACUCCUCUUUGACUUCCAGUCGCCGUCGUCACUAUUGGUGCUAAGCAAAUUUCG-CACUC .............((.(((..((((((((((((..(((((((.......)))))))...)))))))....(((.((....)).))))))))..))).))........-..... ( -30.70, z-score = -4.38, R) >droYak2.chrX 17923162 112 + 21770863 GUCCUUCGCAAACGCCUAGUGCUAAUGUCAAAGUAAGUCAACCGCAUUUGUUGACUCCUCUUUGACUUGCAGUCGCCGUCUUCACUGUUGGUGCUAAGCAAAUUUCG-CACUC .......((....))..(((((....(((((((..(((((((.......)))))))...)))))))((((((.(((((.(......).)))))))..)))).....)-)))). ( -32.50, z-score = -3.44, R) >droEre2.scaffold_4690 12967422 112 - 18748788 CUCCUUCUCAAAUGCCUAGUGCUAAUGUCAAAGUAAGUCAACCGCUUUUGUUGACUCCUCUUUGACUUCCAGUCGCCGUCGUCACUGUUGGUGCCAAGCAAAUUUCG-CACUC ............(((...(..((((.(((((((..(((((((.......)))))))...)))))))...((((.((....)).))))))))..)...))).......-..... ( -28.40, z-score = -3.37, R) >droAna3.scaffold_13334 1361237 95 - 1562580 AUCCUCCUCA--------GCGCUAAUGUUAAACGAAGUCAACCACUUUUGUUGAUUCCCUUUUGACUU----------UCGUCACUUAAAGUGCUAAACAAAUUUGGACACUC .(((.....(--------(((((..........(.(((((((.......))))))).)....((((..----------..)))).....))))))..........)))..... ( -15.16, z-score = -0.97, R) >consensus GUCCUUCUCAAACGCCUAGUGCUAAUGUCAAAGUAAGUCAACCGCUUUUGUUGACUCCUCUUUGACUUCCAGUCGCCGUCGUCACUGUUGGUGCUAAGCAAAUUUCG_CACUC .............((.(((((((((.(((((((..(((((((.......)))))))...)))))))...((((.((....)).))))))))))))).)).............. (-23.02 = -25.13 + 2.11)
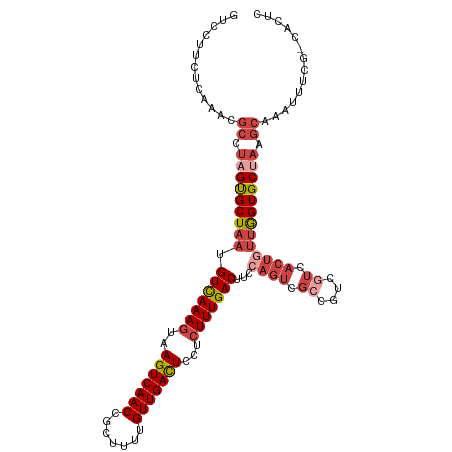
| Location | 9,333,545 – 9,333,657 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 87.00 |
| Shannon entropy | 0.25629 |
| G+C content | 0.45926 |
| Mean single sequence MFE | -30.82 |
| Consensus MFE | -22.06 |
| Energy contribution | -23.42 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.788362 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
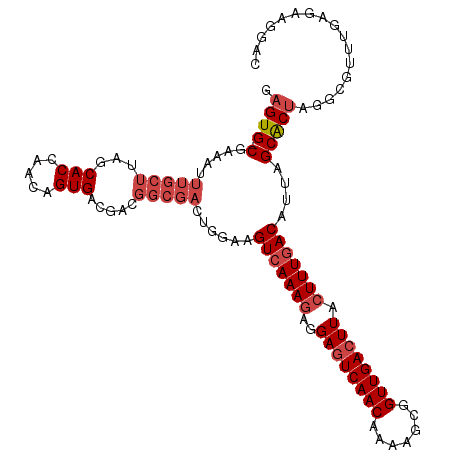
>dm3.chrX 9333545 112 - 22422827 GAGUG-CGAAAUUUGCUUAGCACCAACAGUGACGACGGCGACUGGAAGUCAAAGAGGAGUCAACAAAAGCGGUUGACUUACUUUGACAUUAGCACUACGCGUUUGAGAAGGAC .((((-(.....(((((..((((.....))).)...)))))......(((((((..((((((((.......)))))))).)))))))....)))))................. ( -32.30, z-score = -2.09, R) >droSim1.chrX 7430614 112 - 17042790 GAGUG-CGAAAUUUGCUUAGCACCAACAGUGACGACGGCGACUGGAAGUCAAAGAGGAGUCAACAAAAGCGGUUGACUUACUUUGACAUUAGCACUAGGCGUUUGGGAAGGAC .((((-(.....(((((..((((.....))).)...)))))......(((((((..((((((((.......)))))))).)))))))....)))))................. ( -32.30, z-score = -2.00, R) >droSec1.super_15 102224 112 - 1954846 GAGUG-CGAAAUUUGCUUAGCACCAAUAGUGACGACGGCGACUGGAAGUCAAAGAGGAGUCAACAAAAGCGGUUGACUUACUUUGACAUUAGCACUAGGCGUUUGAGAAGGAC .((((-(.....(((((..((((.....))).)...)))))......(((((((..((((((((.......)))))))).)))))))....)))))................. ( -32.30, z-score = -2.37, R) >droYak2.chrX 17923162 112 - 21770863 GAGUG-CGAAAUUUGCUUAGCACCAACAGUGAAGACGGCGACUGCAAGUCAAAGAGGAGUCAACAAAUGCGGUUGACUUACUUUGACAUUAGCACUAGGCGUUUGCGAAGGAC ...((-((((...((((((((((.....)))...........((((((((((((..((((((((.......)))))))).))))))).)).))))))))))))))))...... ( -32.90, z-score = -1.74, R) >droEre2.scaffold_4690 12967422 112 + 18748788 GAGUG-CGAAAUUUGCUUGGCACCAACAGUGACGACGGCGACUGGAAGUCAAAGAGGAGUCAACAAAAGCGGUUGACUUACUUUGACAUUAGCACUAGGCAUUUGAGAAGGAG .((((-(.....((((((.((((.....))).).).)))))......(((((((..((((((((.......)))))))).)))))))....)))))................. ( -32.00, z-score = -2.09, R) >droAna3.scaffold_13334 1361237 95 + 1562580 GAGUGUCCAAAUUUGUUUAGCACUUUAAGUGACGA----------AAGUCAAAAGGGAAUCAACAAAAGUGGUUGACUUCGUUUAACAUUAGCGC--------UGAGGAGGAU ....((((.....(.((((((.((.....((((..----------..))))(((.(((.(((((.......))))).))).)))......)).))--------)))).))))) ( -23.10, z-score = -1.54, R) >consensus GAGUG_CGAAAUUUGCUUAGCACCAACAGUGACGACGGCGACUGGAAGUCAAAGAGGAGUCAACAAAAGCGGUUGACUUACUUUGACAUUAGCACUAGGCGUUUGAGAAGGAC .((((.(.....(((((...(((.....))).....)))))......(((((((..((((((((.......)))))))).)))))))....)))))................. (-22.06 = -23.42 + 1.36)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:28:15 2011