| Sequence ID | dm3.chrX |
|---|---|
| Location | 9,326,670 – 9,326,772 |
| Length | 102 |
| Max. P | 0.946672 |

| Location | 9,326,670 – 9,326,772 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 66.96 |
| Shannon entropy | 0.62326 |
| G+C content | 0.32111 |
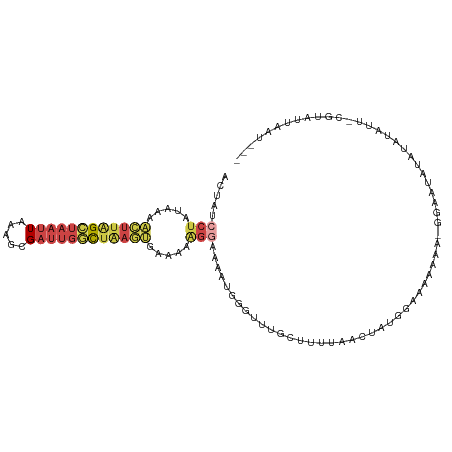
| Mean single sequence MFE | -20.48 |
| Consensus MFE | -9.11 |
| Energy contribution | -10.20 |
| Covariance contribution | 1.09 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.946672 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
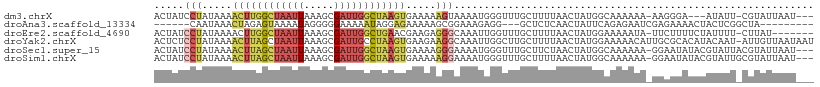
>dm3.chrX 9326670 102 - 22422827 ACUAUCCUAUAAAACUUGGCUAAUUAAAGCGAUUGGCUAAGUGAAAAAGUAAAAUGGGUUUGCUUUUAACUAUGGCAAAAAA-AAGGGA---AUAUU-CGUAUUAAU--- ....((((.....((((((((((((.....))))))))))))..(((((((((.....)))))))))...............-..))))---.....-.........--- ( -22.90, z-score = -2.77, R) >droAna3.scaffold_13334 1354734 92 + 1562580 ------CAAUAAACUAGAGUAAAAUAGGGGGAAAAAUAGGAGAAAAAGCGGAAAGAGG---GCUCUCAACUAUUCAGAGAAUCGAGAAAACUACUCGGCUA--------- ------.......(((........)))...................(((.((...((.---..((((....((((...)))).))))...))..)).))).--------- ( -10.00, z-score = 1.03, R) >droEre2.scaffold_4690 12960752 101 + 18748788 ACUAUCCUAUAAAACUUGGCUAAUUAAAGCGAUUGGCUGAACGAAGAGGGCAAAUUGGUUUGCUUUUAACUAUGGAAAAAUA-UUCUUUUCUAUUUU-CUUAU------- ...............(..(((((((.....)))))))..)..((((((((((((....)))))))))....((((((((...-...)))))))).))-)....------- ( -20.60, z-score = -1.95, R) >droYak2.chrX 17916203 109 - 21770863 ACUCUCCUAUAAAACUUAGCUAAUUAAAGCGAUUGCCUAAGUGAAGAAGGCAAAUUGGCUUGCUUUUAACUAUGGAAAAACAUUGCGCACAUACAAU-AUUGUUAAUAAU ......(((((..((((((.(((((.....))))).))))))...((((((((......))))))))...)))))......((((........))))-............ ( -16.70, z-score = 0.37, R) >droSec1.super_15 95588 106 - 1954846 ACUAUCCUAUAAAACUUAGCUAAUUAAAGCGAUUGGCUAAGUGAAAAGGGAAAAUGGGUUUGCUUCUAACUAUGGCAAAAAA-GGAAUAUACGUAUUACGUAUUAAU--- .(((((((.....((((((((((((.....)))))))))))).....))))...))).((((((.........))))))...-.....(((((.....)))))....--- ( -26.20, z-score = -3.47, R) >droSim1.chrX 7423780 106 - 17042790 ACUAUCCUAUAAAACUUAGCUAAUUAAAGCGAUUGGCUAAGUGAAAAAGGAAAAUGGGUUUGCUUUUAACUAUGGCAAAAAA-GGAAUAUACGUAUUGCGUAUUAAU--- .(((((((.....((((((((((((.....)))))))))))).....))))...))).((((((.........))))))...-.....(((((.....)))))....--- ( -26.50, z-score = -3.47, R) >consensus ACUAUCCUAUAAAACUUAGCUAAUUAAAGCGAUUGGCUAAGUGAAAAAGGAAAAUGGGUUUGCUUUUAACUAUGGAAAAAAA_GGAAUAUAUAUAUU_CGUAUUAAU___ .....(((.....((((((((((((.....)))))))))))).....)))............................................................ ( -9.11 = -10.20 + 1.09)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:28:10 2011