| Sequence ID | dm3.chr2L |
|---|---|
| Location | 10,748,194 – 10,748,286 |
| Length | 92 |
| Max. P | 0.999280 |

| Location | 10,748,194 – 10,748,286 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 68.46 |
| Shannon entropy | 0.54249 |
| G+C content | 0.50191 |
| Mean single sequence MFE | -34.52 |
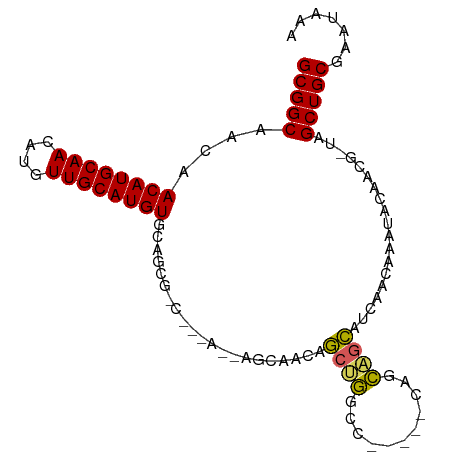
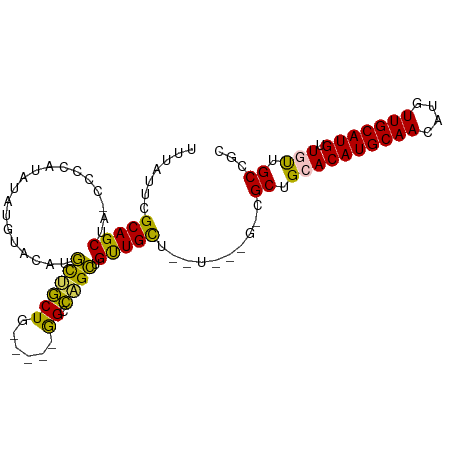
| Consensus MFE | -21.16 |
| Energy contribution | -20.36 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.76 |
| SVM RNA-class probability | 0.999280 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
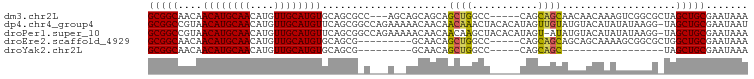
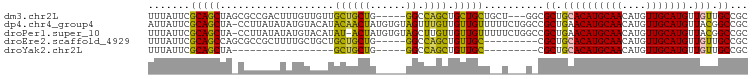
>dm3.chr2L 10748194 92 + 23011544 GCGGCAACAACAUGCAACAUGUUGCAUGUGCAGCGCC---AGCAGCAGCAGCUGGCC-----CAGCAGCAACAACAAAGUCGGCGCUAGCUGCGAAUAAA (((((....((((((((....))))))))..((((((---.((....)).((((...-----...))))............)))))).)))))....... ( -38.80, z-score = -2.05, R) >dp4.chr4_group4 2952874 99 + 6586962 GCGGCCGUAACAUGCAACAUGUUGCAUGUUCAGCGGCCAGAAAAACAACAACAAACUACACAUAGUUGUAUGUACAUAUAUAAGG-UAGCUGCGAAUAAU (((((((((((((((((....)))))))))..))))))......((((((((............))))).)))............-.....))....... ( -30.10, z-score = -3.06, R) >droPer1.super_10 1967495 98 + 3432795 GCGGCCGUAACAUGCAACAUGUUGCAUGUUCAGCGGCCAGAAAAACAACAACAAGCUACACAUAGU-AUAUGUACAUAUAUAAGG-UAGCUGCGAAUAAA (((((((((((((((((....)))))))))..))))))...............((((((...(.((-((((....)))))).).)-)))))))....... ( -36.50, z-score = -5.51, R) >droEre2.scaffold_4929 11958144 86 - 26641161 GCGGCAACAACAUGCAACAUGUUGCAUGUGCAGCG---------GCAACAGCUGGCC-----CAGCAGCAGCAGCAAAAGCGGCGCUGGCUGCGAAUAAA (((((....((((((((....)))))))).(((((---------.(....((((...-----...))))....((....))).))))))))))....... ( -37.10, z-score = -1.16, R) >droYak2.chr2L 7152858 69 + 22324452 GCGGCAACAACAUGCAACAUGUUGCAUGUGCAGCG---------GCAACAGCUGGCC-----CAGCAGC-----------------UAGCUGCGAAUAAA ((.((....((((((((....))))))))...)).---------))..((((((((.-----.....))-----------------))))))........ ( -30.10, z-score = -2.33, R) >consensus GCGGCAACAACAUGCAACAUGUUGCAUGUGCAGCG_C___A__AGCAACAGCUGGCC_____CAGCAGCAUCAACAAAUACAACG_UAGCUGCGAAUAAA (((((....((((((((....)))))))).....................((((.(........)))))...................)))))....... (-21.16 = -20.36 + -0.80)
| Location | 10,748,194 – 10,748,286 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 68.46 |
| Shannon entropy | 0.54249 |
| G+C content | 0.50191 |
| Mean single sequence MFE | -32.80 |
| Consensus MFE | -19.67 |
| Energy contribution | -18.03 |
| Covariance contribution | -1.64 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.95 |
| SVM RNA-class probability | 0.976497 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 10748194 92 - 23011544 UUUAUUCGCAGCUAGCGCCGACUUUGUUGUUGCUGCUG-----GGCCAGCUGCUGCUGCU---GGCGCUGCACAUGCAACAUGUUGCAUGUUGUUGCCGC .......((((((((((.((((......)))).)))))-----.((((((.......)))---))))))))((((((((....))))))))......... ( -40.40, z-score = -1.59, R) >dp4.chr4_group4 2952874 99 - 6586962 AUUAUUCGCAGCUA-CCUUAUAUAUGUACAUACAACUAUGUGUAGUUUGUUGUUGUUUUUCUGGCCGCUGAACAUGCAACAUGUUGCAUGUUACGGCCGC .......(((((.(-(....((((((((........))))))))....)).)))))......(((((...(((((((((....))))))))).))))).. ( -31.40, z-score = -2.79, R) >droPer1.super_10 1967495 98 - 3432795 UUUAUUCGCAGCUA-CCUUAUAUAUGUACAUAU-ACUAUGUGUAGCUUGUUGUUGUUUUUCUGGCCGCUGAACAUGCAACAUGUUGCAUGUUACGGCCGC .......(((((((-((.(((((((....))))-).)).).))))))...............(((((...(((((((((....))))))))).))))))) ( -32.00, z-score = -3.12, R) >droEre2.scaffold_4929 11958144 86 + 26641161 UUUAUUCGCAGCCAGCGCCGCUUUUGCUGCUGCUGCUG-----GGCCAGCUGUUGC---------CGCUGCACAUGCAACAUGUUGCAUGUUGUUGCCGC .......(((((((((((.((.......)).)).))))-----(((........))---------))))))((((((((....))))))))......... ( -33.10, z-score = 0.02, R) >droYak2.chr2L 7152858 69 - 22324452 UUUAUUCGCAGCUA-----------------GCUGCUG-----GGCCAGCUGUUGC---------CGCUGCACAUGCAACAUGUUGCAUGUUGUUGCCGC .......((((((.-----------------(((....-----))).)))))).((---------.((.((((((((((....))))))).))).)).)) ( -27.10, z-score = -0.84, R) >consensus UUUAUUCGCAGCUA_CCCCAUAUAUGUACAUGCUGCUG_____GGCCAGCUGUUGCU__U___G_CGCUGCACAUGCAACAUGUUGCAUGUUGUUGCCGC .......(((((...................((((((......)).)))).)))))..........((.((((((((((....))))))).))).))... (-19.67 = -18.03 + -1.64)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:30:51 2011