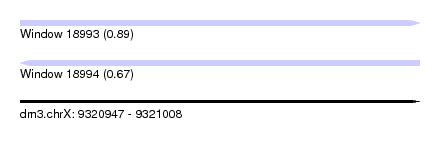
| Sequence ID | dm3.chrX |
|---|---|
| Location | 9,320,947 – 9,321,008 |
| Length | 61 |
| Max. P | 0.893198 |

| Location | 9,320,947 – 9,321,008 |
|---|---|
| Length | 61 |
| Sequences | 8 |
| Columns | 64 |
| Reading direction | forward |
| Mean pairwise identity | 59.34 |
| Shannon entropy | 0.81423 |
| G+C content | 0.51593 |
| Mean single sequence MFE | -14.50 |
| Consensus MFE | -3.99 |
| Energy contribution | -3.29 |
| Covariance contribution | -0.70 |
| Combinations/Pair | 1.92 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.28 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.893198 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
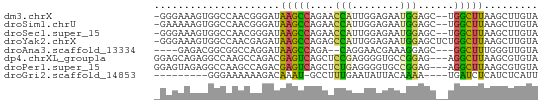
>dm3.chrX 9320947 61 + 22422827 UACAAGCUUAAGCCA--GCUCCAUUCUCCAAUGGUUCUGGCUUAUCCCGUUGGCCACUUUCCC- .....((((((((((--(..(((((....)))))..)))))))).......))).........- ( -17.71, z-score = -3.51, R) >droSim1.chrU 3390 61 - 15797150 UACAAGCUUAAGCCA--GCUCCAUUCUCCAAUGGUUCUGGCUUAUCCCGUUGGCCACUUUUUC- .....((((((((((--(..(((((....)))))..)))))))).......))).........- ( -17.71, z-score = -3.61, R) >droSec1.super_15 89991 61 + 1954846 UACAAGCUUAAGCCA--GCUCCAUUCUCCAAUGGUUCUGGCUUAUCCCGUUGGCCACUUUCCC- .....((((((((((--(..(((((....)))))..)))))))).......))).........- ( -17.71, z-score = -3.51, R) >droYak2.chrX 17909357 63 + 21770863 UACAAGCUUAAGCCAGAGCUCCAUUCUCCAAUGGCUCUGGCUUAUCUCGUUGGCCACUUUCCC- .....((((((((((((((..((((....))))))))))))))).......))).........- ( -19.81, z-score = -3.32, R) >droAna3.scaffold_13334 1349367 55 - 1562580 UACAACCCAAAGCC---GCUCCUUUCGUUCCUG--UCUGGCUUAUCCUGGCCGCCGUCUC---- ......((((((((---(..(...........)--..))))))....)))..........---- ( -6.00, z-score = 0.91, R) >dp4.chrXL_group1a 1472423 61 - 9151740 UACACGCUUAAGCCU---CUCCGGCACCCCUCGGAGCUGACUCGUCUGGCUUGGCCUCUGCUCC .....(((.(((((.---((((((......))))))..((....)).))))))))......... ( -16.60, z-score = -0.45, R) >droPer1.super_15 2159041 61 - 2181545 UACACGCUUAAGCCU---CUCCGGCACCCCUCAGAGCUGACUCGUCUGGCUUGGCCUCUACUCC .....(((.(((((.---....((....))...(((....)))....))))))))......... ( -11.90, z-score = 0.44, R) >droGri2.scaffold_14853 804908 50 - 10151454 AAUGAGAUGAGAUCA----UUUUGUAAUAUUCAAAGGC-AUUUGUCUUUUUUCCC--------- .(..(((((....))----)))..).......((((((-....))))))......--------- ( -8.60, z-score = -1.04, R) >consensus UACAAGCUUAAGCCA__GCUCCAUUCUCCAAUGGAUCUGGCUUAUCCCGUUGGCCACUUUCCC_ .....((.((((((......((((......))))....))))))....)).............. ( -3.99 = -3.29 + -0.70)


| Location | 9,320,947 – 9,321,008 |
|---|---|
| Length | 61 |
| Sequences | 8 |
| Columns | 64 |
| Reading direction | reverse |
| Mean pairwise identity | 59.34 |
| Shannon entropy | 0.81423 |
| G+C content | 0.51593 |
| Mean single sequence MFE | -18.05 |
| Consensus MFE | -4.41 |
| Energy contribution | -3.55 |
| Covariance contribution | -0.86 |
| Combinations/Pair | 2.00 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.24 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.666944 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chrX 9320947 61 - 22422827 -GGGAAAGUGGCCAACGGGAUAAGCCAGAACCAUUGGAGAAUGGAGC--UGGCUUAAGCUUGUA -((........)).(((((.((((((((..(((((....)))))..)--)))))))..))))). ( -22.70, z-score = -3.38, R) >droSim1.chrU 3390 61 + 15797150 -GAAAAAGUGGCCAACGGGAUAAGCCAGAACCAUUGGAGAAUGGAGC--UGGCUUAAGCUUGUA -.............(((((.((((((((..(((((....)))))..)--)))))))..))))). ( -21.90, z-score = -3.60, R) >droSec1.super_15 89991 61 - 1954846 -GGGAAAGUGGCCAACGGGAUAAGCCAGAACCAUUGGAGAAUGGAGC--UGGCUUAAGCUUGUA -((........)).(((((.((((((((..(((((....)))))..)--)))))))..))))). ( -22.70, z-score = -3.38, R) >droYak2.chrX 17909357 63 - 21770863 -GGGAAAGUGGCCAACGAGAUAAGCCAGAGCCAUUGGAGAAUGGAGCUCUGGCUUAAGCUUGUA -((........)).(((((.(((((((((((((((....))))..)))))))))))..))))). ( -25.40, z-score = -3.63, R) >droAna3.scaffold_13334 1349367 55 + 1562580 ----GAGACGGCGGCCAGGAUAAGCCAGA--CAGGAACGAAAGGAGC---GGCUUUGGGUUGUA ----......((((((.....(((((.(.--......(....)...)---)))))..)))))). ( -13.80, z-score = -0.81, R) >dp4.chrXL_group1a 1472423 61 + 9151740 GGAGCAGAGGCCAAGCCAGACGAGUCAGCUCCGAGGGGUGCCGGAG---AGGCUUAAGCGUGUA ...((..(((((......((....))..(((((........)))))---.)))))..))..... ( -18.60, z-score = 0.11, R) >droPer1.super_15 2159041 61 + 2181545 GGAGUAGAGGCCAAGCCAGACGAGUCAGCUCUGAGGGGUGCCGGAG---AGGCUUAAGCGUGUA ...((..(((((...((....(((....)))...((....))))..---.)))))..))..... ( -15.50, z-score = 0.87, R) >droGri2.scaffold_14853 804908 50 + 10151454 ---------GGGAAAAAAGACAAAU-GCCUUUGAAUAUUACAAAA----UGAUCUCAUCUCAUU ---------((((..((((......-..))))((.((((....))----)).))...))))... ( -3.80, z-score = 0.50, R) >consensus _GAGAAAGUGGCCAACGAGAUAAGCCAGAACCAUUGGAGAAUGGAGC__UGGCUUAAGCUUGUA .....................(((((....(((........)))......)))))......... ( -4.41 = -3.55 + -0.86)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:28:09 2011