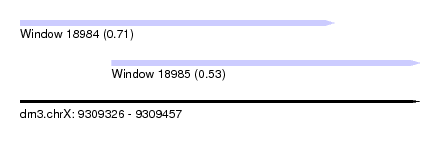
| Sequence ID | dm3.chrX |
|---|---|
| Location | 9,309,326 – 9,309,457 |
| Length | 131 |
| Max. P | 0.709644 |

| Location | 9,309,326 – 9,309,429 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 79.36 |
| Shannon entropy | 0.34070 |
| G+C content | 0.47268 |
| Mean single sequence MFE | -16.05 |
| Consensus MFE | -11.09 |
| Energy contribution | -11.73 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.709644 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
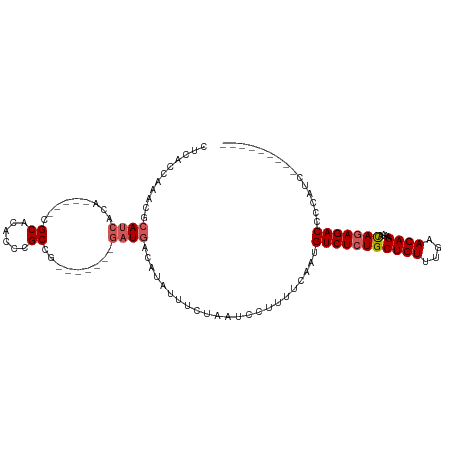
>dm3.chrX 9309326 103 + 22422827 UUCAACAAACGCAACACA-----CGCACACCCGCCG-------GAUGACAUAUUUCUAAUCCUUUUCAAUGUCUCUGCUGUUUGAACAGAAAACAGAGACCCCCCCGUUCCGACC .........((.(((...-----.((......)).(-------(((............))))........(((((((((((....))))....)))))))......))).))... ( -18.20, z-score = -2.00, R) >droEre2.scaffold_4690 12942225 80 - 18748788 ------------AUCACA-----CGCACACCCGCCG-------UAUGACACAUUUCUAAUCCUUUUCAAUGUCUCUGCUGUUUGAACAGCAUAUAGAGACCCCA----------- ------------.(((.(-----(((......).))-------).)))......................(((((((((((....)))))....))))))....----------- ( -15.80, z-score = -2.85, R) >droYak2.chrX 17898042 106 + 21770863 CUCACAAAACGCAUCACACAUCACGCACACCCGCCGCCGGACGGAUGACAUAUUUCUAAUCCUUUUCAAUGUCUCUGCUGUUUGAACAGAAAACAAAGACCCCAGC--------- ........................((......)).((.((.((((.(((((.................)))))))))((((....))))............)).))--------- ( -12.73, z-score = 0.95, R) >droSec1.super_15 79057 94 + 1954846 CUCACCAAACGCAUCA-------CGCACACCCGCCG-------GAUGACAUAUUUCUAAUCCUUUUCAAUGUCUCUGCUGUUUGAACAGAAAACAGAGACCCCUUCCG------- ..........(((((.-------.((......))..-------)))).).....................(((((((((((....))))....)))))))........------- ( -16.80, z-score = -2.08, R) >droSim1.chrX 7409166 94 + 17042790 CUCACCAAAAGCAUCA-------CGCACACCCGCCG-------GAUGACAUAUUUCUAAUCCUUUUCAAUGUCUCUGCUGUUUGAACAGAAAACAGAGACCCCUUCCG------- ..........(((((.-------.((......))..-------)))).).....................(((((((((((....))))....)))))))........------- ( -16.70, z-score = -1.89, R) >consensus CUCACCAAACGCAUCACA_____CGCACACCCGCCG_______GAUGACAUAUUUCUAAUCCUUUUCAAUGUCUCUGCUGUUUGAACAGAAAACAGAGACCCCAUC_________ ...........((((.........((......)).........)))).......................(((((((((((....))))....)))))))............... (-11.09 = -11.73 + 0.64)

| Location | 9,309,356 – 9,309,457 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 76.87 |
| Shannon entropy | 0.38696 |
| G+C content | 0.44951 |
| Mean single sequence MFE | -16.21 |
| Consensus MFE | -11.76 |
| Energy contribution | -12.00 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.02 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.528035 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
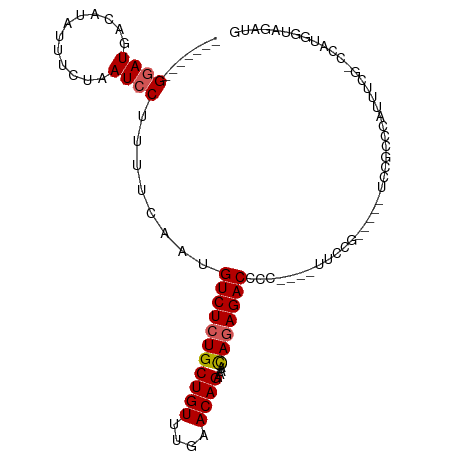
>dm3.chrX 9309356 101 + 22422827 -------GGAUGACAUAUUUCUAAUCCUUUUCAAUGUCUCUGCUGUUUGAACAGAAAACAGAGACCCCCCCGUUCCGACCCUUCCUCCCGUUUCGCCCUUGGUAGAUG -------((((............))))........(((((((((((....))))....))))))).(..(((...(((..(........)..)))....)))..)... ( -17.70, z-score = -0.84, R) >droEre2.scaffold_4690 12942243 78 - 18748788 -------GUAUGACACAUUUCUAAUCCUUUUCAAUGUCUCUGCUGUUUGAACAGCAUAUAGAGACCCC---AUUCCG------CCCGCAAUAUA-------------- -------............................(((((((((((....)))))....))))))...---......------...........-------------- ( -12.40, z-score = -1.74, R) >droYak2.chrX 17898077 96 + 21770863 GCCGGACGGAUGACAUAUUUCUAAUCCUUUUCAAUGUCUCUGCUGUUUGAACAGAAAACAAAGACCCC-----AGCG-----UCCGCUCAUAUA--CCAUACUAUAUA ((.(((((...(((((.................)))))(((..(((((.......))))).)))....-----..))-----)))))..(((((--......))))). ( -17.03, z-score = -1.20, R) >droSec1.super_15 79085 93 + 1954846 -------GGAUGACAUAUUUCUAAUCCUUUUCAAUGUCUCUGCUGUUUGAACAGAAAACAGAGACCCC----UUCCG----CUCCGCCCAUUCCGCCCAUGGUAGAUG -------((((............))))........(((((((((((....))))....)))))))...----.....----..........((..((...))..)).. ( -16.50, z-score = -0.59, R) >droSim1.chrX 7409194 93 + 17042790 -------GGAUGACAUAUUUCUAAUCCUUUUCAAUGUCUCUGCUGUUUGAACAGAAAACAGAGACCCC----UUCCG----CUCCGCCCAUUCCGUCCAUGGUAGAUG -------(((((.......................(((((((((((....))))....)))))))...----....(----(...))......))))).......... ( -17.40, z-score = -0.75, R) >consensus _______GGAUGACAUAUUUCUAAUCCUUUUCAAUGUCUCUGCUGUUUGAACAGAAAACAGAGACCCC____UUCCG_____UCCGCCCAUUUCG_CCAUGGUAGAUG .......((((............))))........(((((((((((....))))....)))))))........................................... (-11.76 = -12.00 + 0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:28:01 2011