| Sequence ID | dm3.chrX |
|---|---|
| Location | 9,281,264 – 9,281,373 |
| Length | 109 |
| Max. P | 0.845410 |

| Location | 9,281,264 – 9,281,373 |
|---|---|
| Length | 109 |
| Sequences | 11 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 71.05 |
| Shannon entropy | 0.58367 |
| G+C content | 0.38105 |
| Mean single sequence MFE | -23.62 |
| Consensus MFE | -10.67 |
| Energy contribution | -10.67 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.845410 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
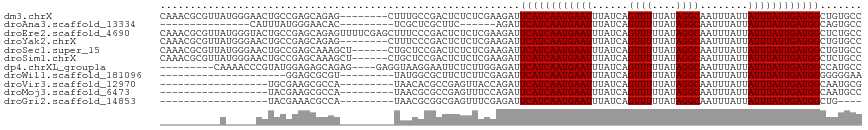
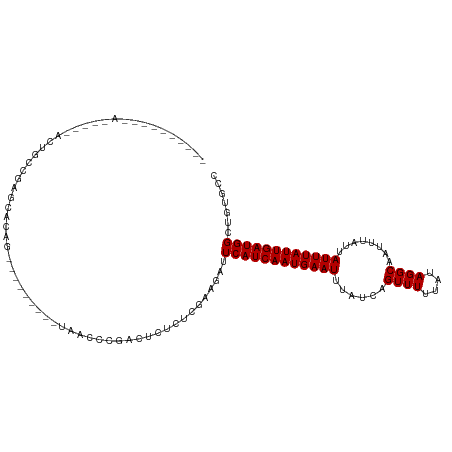
>dm3.chrX 9281264 109 + 22422827 CAAACGCGUUAUGGGAACUGCCGAGCAGAG--------CUUUGCCGACUCUCUCGAAGAUUCAUCAAUGAAUUUAUCAGUUUUUAUAGGCAAUUUAUUAUUUAUUGAUGGCUGUGCC .........(((((((((((.((((.((((--------(......).)))))))).(((((((....)))))))..)))))))))))((((..((((((.....))))))...)))) ( -28.90, z-score = -1.73, R) >droAna3.scaffold_13334 1307869 87 - 1562580 ---------------CAUUUAUGGGAACAC---------UCGCUCGCUUC------AGAUUCAUCAAUGAAUUUAUCAGUUUUUAUAGGCAAUUUAUUAUUUAUUGAUGGCAGUGCC ---------------............(((---------(.((((.....------.))..(((((((((((......((((....))))........))))))))))))))))).. ( -15.34, z-score = -0.60, R) >droEre2.scaffold_4690 12912015 117 - 18748788 CAAACGCGUUAUGGGUACUGCCGAGCAGAGUUUUCGAGCUUUCCCGACUCUCUCGAAGAUUCAUCAAUGAAUUUAUCAGUUUUUAUAGGCAAUUUAUUAUUUAUUGAUGGCUCUGCC .....((......((.....))((((.(((((((((((.............)))))))))))((((((((((......((((....))))........)))))))))).)))).)). ( -29.16, z-score = -1.59, R) >droYak2.chrX 17873230 109 + 21770863 CAAACGCGUUAUGGGAACUGCCGAGCAGAG--------CUUUCCCGACUCUCUCGAAGAUUCAUCAAUGAAUUUAUCAGUUUUUAUAGGCAAUUUAUUAUUUAUUGAUGGCUGUGCC .........(((((((((((.((((.((((--------(......).)))))))).(((((((....)))))))..)))))))))))((((..((((((.....))))))...)))) ( -28.90, z-score = -1.98, R) >droSec1.super_15 50592 111 + 1954846 CAAACGCGUUAUGGGAACUGCCGAGCAAAGCU------CUGCUCCGACUCUCUCGAAGAUUCAUCAAUGAAUUUAUCAGUUUUUAUAGGCAAUUUAUUAUUUAUUGAUGGCUGUGCC .........(((((((((((..(((((.....------.)))))(((.....))).(((((((....)))))))..)))))))))))((((..((((((.....))))))...)))) ( -26.40, z-score = -0.97, R) >droSim1.chrX 7392350 111 + 17042790 CAAACGCGUUAUGGGAACUGCCGAGCAAAGCU------CUGCUCCGACUCUCUCGAAGAUUCAUCAAUGAAUUUAUCAGUUUUUAUAGGCAAUUUAUUAUUUAUUGAUGGCUCUGCC .....((....(((((....(.(((((.....------.))))).)....))))).(((..(((((((((((......((((....))))........)))))))))))..))))). ( -26.24, z-score = -1.11, R) >dp4.chrXL_group1a 1431695 104 - 9151740 ---------CAAAACCCGUAUGGAGAGCAGAG----GAGGUAAGGAAUUCUCUUGGAGAUUCAUCAAUGAAUUUAUCAGUUUUUAUAGGCAAUUUAUUAUUUAUUGAUGGCCAUGCC ---------........((((((.......((----((((........))))))......((((((((((((......((((....))))........)))))))))))))))))). ( -23.24, z-score = -0.82, R) >droWil1.scaffold_181096 2783182 87 + 12416693 ---------------------GGAGCGCGU---------UAUGGCGCUUCUCUUCGAGAUUCAUCAAUGAAUUUAUCAGUUUUUAUAGGCAAUUUAUUAUUUAUUGAUGGGGGGGAA ---------------------(((((((..---------....)))))))(((((....(((((((((((((......((((....))))........)))))))))))))))))). ( -25.24, z-score = -3.39, R) >droVir3.scaffold_12970 6012304 90 - 11907090 ------------------UGCGAAGCGCCA---------UAACACGCCGAGUUACCAGAUUCAUCAAUGAAUUUAUCAGUUUUUAUAGGCAAUUUAUUAUUUAUUGAUGGCAAUGCG ------------------.(((...)))..---------......((((((((....)))))((((((((((......((((....))))........)))))))))))))...... ( -16.64, z-score = -0.52, R) >droMoj3.scaffold_6473 15732676 90 + 16943266 ------------------UACGAAGCGCCA---------UAACGCGCCGAGUUUCCAGAUUCAUCAAUGAAUUUAUCAGUUUUUAUAGGCAAUUUAUUAUUUAUUGAUGGCAAUGCC ------------------......(((...---------...)))((((((((....)))))((((((((((......((((....))))........)))))))))))))...... ( -19.24, z-score = -1.61, R) >droGri2.scaffold_14853 9388373 86 + 10151454 ------------------UACGAAACGCCA---------UAACGCGGCGAGUUUCGAGAUUCAUCAAUGAAUUUAUCAGUUUUUAUAGGCAAUUUAUUAUUUAUUGAUGGCUG---- ------------------..(((((((((.---------......)))..))))))....((((((((((((......((((....))))........))))))))))))...---- ( -20.54, z-score = -2.37, R) >consensus __________A_____ACUGCCGAGCACAG_________UAACCCGACUCUCUCGAAGAUUCAUCAAUGAAUUUAUCAGUUUUUAUAGGCAAUUUAUUAUUUAUUGAUGGCUGUGCC ............................................................((((((((((((......((((....))))........))))))))))))....... (-10.67 = -10.67 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:27:57 2011