| Sequence ID | dm3.chrX |
|---|---|
| Location | 9,273,691 – 9,273,784 |
| Length | 93 |
| Max. P | 0.836379 |

| Location | 9,273,691 – 9,273,784 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 65.15 |
| Shannon entropy | 0.56870 |
| G+C content | 0.36361 |
| Mean single sequence MFE | -15.04 |
| Consensus MFE | -8.03 |
| Energy contribution | -8.87 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.836379 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
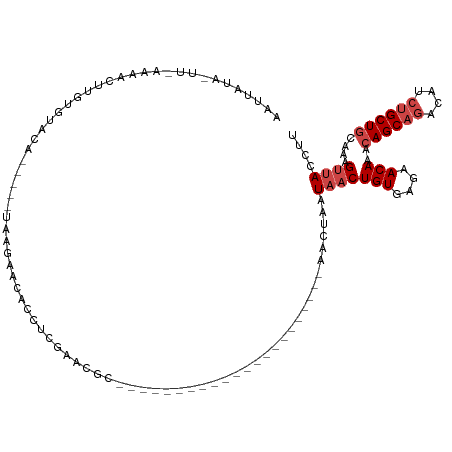
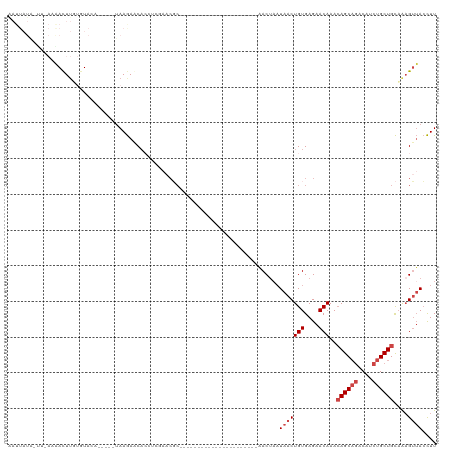
>dm3.chrX 9273691 93 - 22422827 AAUUAUAAUUUAAGAUUUGUUUACA-----UAAGACCACCGCCAACGC----------------------AACUAAUAACUGUGAGAACAAACAGCAGACAUCUGCUGCAAAGUUACCUU .....((((((....(((((((.((-----((.(.....)((....))----------------------..........)))).)))))))((((((....))))))..)))))).... ( -16.80, z-score = -2.43, R) >droSim1.chrX 7385242 78 - 17042790 -------------AAUUUUAAUU-------UAAGAACACCUCGAACGC----------------------AACUAAUAACUGUGAGAACAAAAAGCAGACAUCUGCUGCAAAGUUACCUU -------------((((((..((-------..((.....))..)).((----------------------..........(((....)))...(((((....)))))))))))))..... ( -10.20, z-score = -0.80, R) >droSec1.super_15 43003 78 - 1954846 -------------AAUUAUAAUU-------UAAGAACACCUCGAACGC----------------------AACUAAUAACUGUGAGAACAAACAGCAGACAUCUGCUGCAAAGUUACCUU -------------.....(((((-------(..............(((----------------------(.........))))........((((((....))))))..)))))).... ( -12.20, z-score = -1.81, R) >droYak2.chrX 17865417 81 - 21770863 AAUUAUAAUUUACGACUUUUGUACA-----UAAGAGC----------------------------------ACCUUUAACUGUGAGAACAAACAGCAGACAUCUGCUGCAAAGUUACCUU .............(.((((((....-----)))))))----------------------------------.....(((((((....))...((((((....))))))...))))).... ( -13.70, z-score = -1.35, R) >droEre2.scaffold_4690 12904465 98 + 18748788 AAUUAUAGCCCAAGACUUGUGUACAUAUAGUAAGAGCAAAUACAACGC----------------------AACUAAUAACUGUGAGAACAAACAGCAGACAUCUGCUGCAAAGUUACCUU .......((.......((((.(((.....)))...)))).......))----------------------......(((((((....))...((((((....))))))...))))).... ( -16.34, z-score = -0.35, R) >droAna3.scaffold_13334 1296189 115 + 1562580 GAUUAUGUUUAAAAACUUGUGUACU-----CAAAAACACCUCGAACUCCUCAUCCCGGAGCCAAGCACUUCACUAAUAACUGUGAGAACAAACAGCCAGCAUCUGCUGUUGUGAGAAUUU ..................((((...-----.....))))(((...((((.......))))........(((((........))))).....(((((.(((....)))))))))))..... ( -21.00, z-score = -0.55, R) >consensus AAUUAUA_UU_AAAACUUGUGUACA_____UAAGAACACCUCGAACGC______________________AACUAAUAACUGUGAGAACAAACAGCAGACAUCUGCUGCAAAGUUACCUU ............................................................................(((((((....)))..((((((....))))))....)))).... ( -8.03 = -8.87 + 0.83)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:27:57 2011