
| Sequence ID | dm3.chrX |
|---|---|
| Location | 9,247,230 – 9,247,356 |
| Length | 126 |
| Max. P | 0.860987 |

| Location | 9,247,230 – 9,247,332 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 82.55 |
| Shannon entropy | 0.33330 |
| G+C content | 0.44218 |
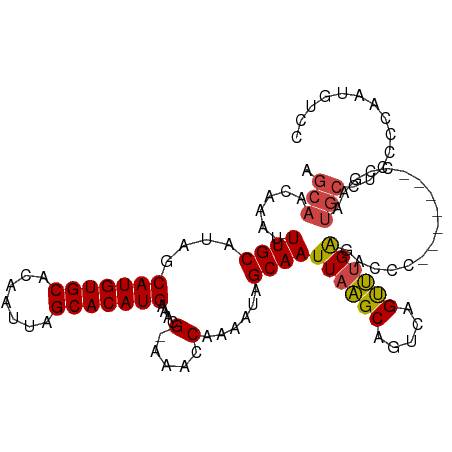
| Mean single sequence MFE | -19.57 |
| Consensus MFE | -14.99 |
| Energy contribution | -14.63 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.860987 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 9247230 102 - 22422827 AGCAACAAAUUUGCAUAGCAUGUGCACAAUUAGCACAUGAAACG-AAACCAAAAUAGCAAUUAAGCAGUCUGUUUGAGACCC-------CCGAAUGCCUCCCCACUGUCC .((((.....))))(((((((((((.......)))))))...((-...........((......)).((((.....))))..-------.))............)))).. ( -18.50, z-score = -1.33, R) >droAna3.scaffold_13334 1266909 87 + 1562580 AAUAAUA-AUUUGCAUACCAUGUGCACAAUUAGCACAUGAAACGAAAACCAAAAUAGCAAUUAGGC---CAGCCAGGCAGCCUC----UCCCCAC--------------- .......-..((((....(((((((.......)))))))....(.....)......))))....((---(.....)))......----.......--------------- ( -16.20, z-score = -1.50, R) >droEre2.scaffold_4690 12875770 101 + 18748788 AGCAACAGAUUUGCAUAGCAUGUGCACAAUUAGCACAUGAAACG-AAACCAAAAUAGCAAUUAAGCGGUCAGUUUGAGACC--------CCAAAUGCCUGCCCAAUGCCC .((((.....))))....(((((((.......))))))).....-...........(((.....((((...(((((.....--------.)))))..))))....))).. ( -22.10, z-score = -1.58, R) >droYak2.chrX 17839849 109 - 21770863 AGCAACAAAUUUGCAUAGCAUGUGCACAAUUAGCACAUGAAACG-AAACCAAAAUAGCAAUUAAGCAGUCCGUUUGAGAGCCCCGAAUGCCGAAUGCCUCCCCAGUGUCG .((((.....))))....(((((((.......)))))))...((-(.((.......(((.((..(((.((.(..(....)..).)).)))..))))).......)).))) ( -21.44, z-score = -1.24, R) >droSec1.super_15 16797 102 - 1954846 AGCAACAAAUUUGCAUAGCAUGUGCACAAUUAGCACAUGAAACG-AAACCAAAAUAGCAAUUAAGCAGCCAGUUUGAGACCC-------CCGAAUGCGUCCCCAAUGUCC .(((.((((((.((....(((((((.......))))))).....-...........((......)).)).)))))).(....-------.)...)))............. ( -19.60, z-score = -1.98, R) >droSim1.chrX_random 291030 102 - 5698898 AGCAACAAAUUUGCAUAGCAUGUGCACAAUUAGCACAUGAAACG-AAACCAAAAUAGCAAUUAAGCAGCCAGUUUGAGACCC-------CCGAAUGCGUCCCCAAUGUCC .(((.((((((.((....(((((((.......))))))).....-...........((......)).)).)))))).(....-------.)...)))............. ( -19.60, z-score = -1.98, R) >consensus AGCAACAAAUUUGCAUAGCAUGUGCACAAUUAGCACAUGAAACG_AAACCAAAAUAGCAAUUAAGCAGUCAGUUUGAGACCC_______CCGAAUGCCUCCCCAAUGUCC .((((.....))))....(((((((.......))))))).....................((((((.....))))))................................. (-14.99 = -14.63 + -0.36)

| Location | 9,247,250 – 9,247,356 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 88.01 |
| Shannon entropy | 0.23012 |
| G+C content | 0.45126 |
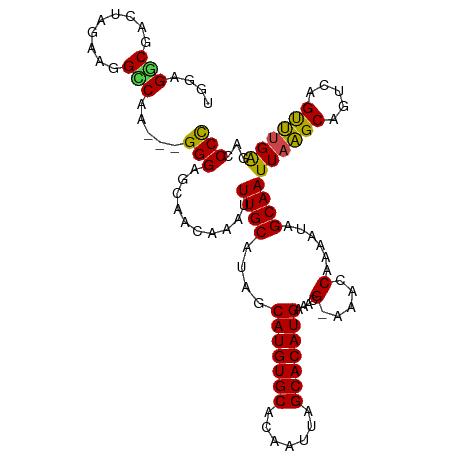
| Mean single sequence MFE | -26.53 |
| Consensus MFE | -20.14 |
| Energy contribution | -19.50 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.647822 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 9247250 106 - 22422827 UUGAGACAACUAGAAGGCCAA---GGGAGCAACAAAUUUGCAUAGCAUGUGCACAAUUAGCACAUGAAACG-AAACCAAAAUAGCAAUUAAGCAGUCUGUUUGAGACCCC ((.((((((((..........---((..((((.....))))....(((((((.......))))))).....-...))......((......))))).))))).))..... ( -21.40, z-score = -1.11, R) >droAna3.scaffold_13334 1266917 106 + 1562580 UGGGGCCUGCUAGGAGGGCUGUCUGGGAAUAAUA-AUUUGCAUACCAUGUGCACAAUUAGCACAUGAAACGAAAACCAAAAUAGCAAUUAGGC---CAGCCAGGCAGCCU ..((((.((((.((..((((((.((.((((....-)))).)).))(((((((.......)))))))........................)))---)..)).)))))))) ( -34.80, z-score = -1.61, R) >droEre2.scaffold_4690 12875789 106 + 18748788 UGGAGGCGACGAGAAGGUCAA---GGGAGCAACAGAUUUGCAUAGCAUGUGCACAAUUAGCACAUGAAACG-AAACCAAAAUAGCAAUUAAGCGGUCAGUUUGAGACCCC .((.(....).....((((..---((..((((.....))))....(((((((.......))))))).....-...))..........((((((.....)))))))))))) ( -25.20, z-score = -1.85, R) >droYak2.chrX 17839876 106 - 21770863 UGGAGGCGACGAGAAGGCCAA---GGGAGCAACAAAUUUGCAUAGCAUGUGCACAAUUAGCACAUGAAACG-AAACCAAAAUAGCAAUUAAGCAGUCCGUUUGAGAGCCC ....(((..((((..(((...---((..((((.....))))....(((((((.......))))))).....-...))......((......)).)))..))))...))). ( -26.10, z-score = -1.98, R) >droSec1.super_15 16817 106 - 1954846 UGGAGGCUACUAGAAGGCCAA---GGGAGCAACAAAUUUGCAUAGCAUGUGCACAAUUAGCACAUGAAACG-AAACCAAAAUAGCAAUUAAGCAGCCAGUUUGAGACCCC ....((((.......))))..---(((..(..((((((.((....(((((((.......))))))).....-...........((......)).)).)))))).)..))) ( -26.60, z-score = -2.08, R) >droSim1.chrX_random 291050 106 - 5698898 UGGAGGCGACUAGAAGGCCAA---GGGAGCAACAAAUUUGCAUAGCAUGUGCACAAUUAGCACAUGAAACG-AAACCAAAAUAGCAAUUAAGCAGCCAGUUUGAGACCCC ....(((.........)))..---(((..(..((((((.((....(((((((.......))))))).....-...........((......)).)).)))))).)..))) ( -25.10, z-score = -1.79, R) >consensus UGGAGGCGACUAGAAGGCCAA___GGGAGCAACAAAUUUGCAUAGCAUGUGCACAAUUAGCACAUGAAACG_AAACCAAAAUAGCAAUUAAGCAGUCAGUUUGAGACCCC ....(((.........))).....(((.((((.....))))....(((((((.......))))))).....................((((((.....))))))...))) (-20.14 = -19.50 + -0.64)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:27:53 2011