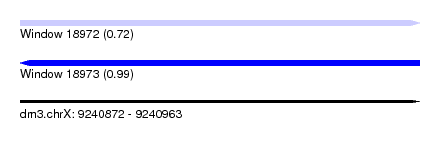
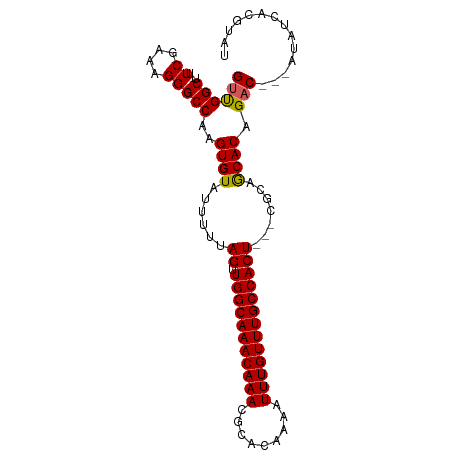
| Sequence ID | dm3.chrX |
|---|---|
| Location | 9,240,872 – 9,240,963 |
| Length | 91 |
| Max. P | 0.990420 |

| Location | 9,240,872 – 9,240,963 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 90.44 |
| Shannon entropy | 0.15788 |
| G+C content | 0.43698 |
| Mean single sequence MFE | -26.25 |
| Consensus MFE | -21.38 |
| Energy contribution | -21.14 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.715160 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
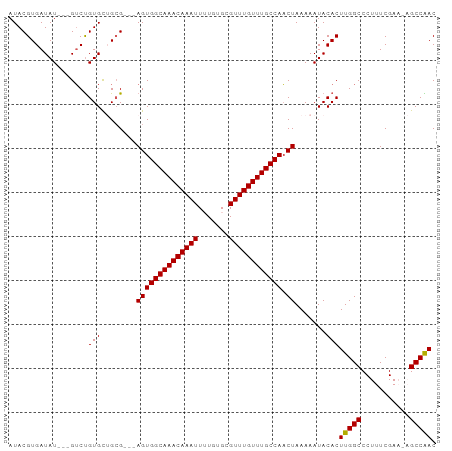
>dm3.chrX 9240872 91 + 22422827 AUACGUGAUAU---GUCUGUGCUGCG---AGUGGCAAACAAAUUUUGUGCGUUUGUUUGCCAACUAAAAAUACACUUGGCCCUUUCGAAUAGCCAAC ...(((..(((---....)))..)))---((((((((((((((.......))))))))))).)))..........(((((...........))))). ( -22.70, z-score = -1.32, R) >droSim1.chrX 7375290 94 + 17042790 AUACGUGAUAUGUUGUCUGUGCUGCG---AGUGGCAAACAAAUUGUGUGCGUUUGUUUGCCAACUAAAAAUACACUUGGCCCUUUCGAACAGCCAAC ...........((((.((((....((---((((((((((((((.......)))))))))))).((((........))))....)))).)))).)))) ( -25.20, z-score = -1.15, R) >droSec1.super_68 130832 94 + 137979 AUACGUGAUAUGUUGUCUGUGCUGAG---AGUGGCAAACAAAUUGUGUGCGUUUGUUUGCCAACUAAAAAUACACUUGGCCCUUUCGAACAGCCAAC ...........((((.((((..((((---((((((((((((((.......)))))))))))).((((........))))..)))))).)))).)))) ( -27.90, z-score = -2.35, R) >droYak2.chrX 17833668 89 + 21770863 AUACGUGAUAU---GUCCGUGUAGCG---AGUGGCAAACAAAUUUUGUGCGUUUGUUUGCCAACUAAAAAUACACUUGGCCCUUUCGAU--GCCGAC ...........---....(((((...---((((((((((((((.......))))))))))).))).....)))))(((((.........--))))). ( -25.60, z-score = -2.23, R) >droEre2.scaffold_4690 12869620 92 - 18748788 AUACGUGAUAU---GGCUGUGUAGCGGCGAGUGGCAAACAAAUUUUGUGCGUUUGUUUGCCACCUAAAAAUACACUUGGCCCUUUCGAU--GCCGAC ...(((((...---(((((((((.......(((((((((((((.......))))))))))))).......)))))..))))...)).))--)..... ( -29.84, z-score = -2.32, R) >consensus AUACGUGAUAU___GUCUGUGCUGCG___AGUGGCAAACAAAUUUUGUGCGUUUGUUUGCCAACUAAAAAUACACUUGGCCCUUUCGAA_AGCCAAC ..................(((........((((((((((((((.......)))))))))))).)).......)))(((((...........))))). (-21.38 = -21.14 + -0.24)

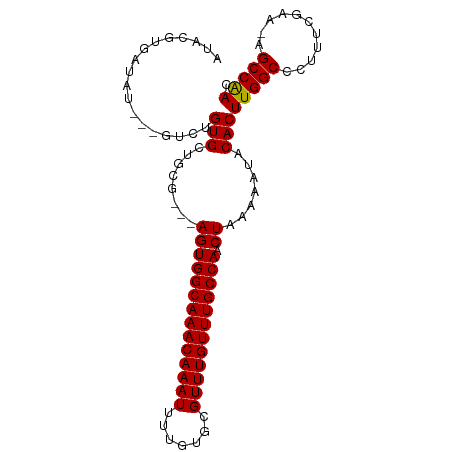
| Location | 9,240,872 – 9,240,963 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 90.44 |
| Shannon entropy | 0.15788 |
| G+C content | 0.43698 |
| Mean single sequence MFE | -27.48 |
| Consensus MFE | -23.78 |
| Energy contribution | -23.50 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.86 |
| Structure conservation index | 0.87 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.42 |
| SVM RNA-class probability | 0.990420 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 9240872 91 - 22422827 GUUGGCUAUUCGAAAGGGCCAAGUGUAUUUUUAGUUGGCAAACAAACGCACAAAAUUUGUUUGCCACU---CGCAGCACAGAC---AUAUCACGUAU .((((((...(....)))))))((((......(((.((((((((((.........)))))))))))))---....))))....---........... ( -25.00, z-score = -2.35, R) >droSim1.chrX 7375290 94 - 17042790 GUUGGCUGUUCGAAAGGGCCAAGUGUAUUUUUAGUUGGCAAACAAACGCACACAAUUUGUUUGCCACU---CGCAGCACAGACAACAUAUCACGUAU ((((.((((((....))))...((((......(((.((((((((((.........)))))))))))))---....)))))).))))........... ( -26.20, z-score = -1.93, R) >droSec1.super_68 130832 94 - 137979 GUUGGCUGUUCGAAAGGGCCAAGUGUAUUUUUAGUUGGCAAACAAACGCACACAAUUUGUUUGCCACU---CUCAGCACAGACAACAUAUCACGUAU ((((.((((((....))))...((((......(((.((((((((((.........)))))))))))))---....)))))).))))........... ( -26.20, z-score = -2.34, R) >droYak2.chrX 17833668 89 - 21770863 GUCGGC--AUCGAAAGGGCCAAGUGUAUUUUUAGUUGGCAAACAAACGCACAAAAUUUGUUUGCCACU---CGCUACACGGAC---AUAUCACGUAU ((((((--.((....)))))..(((((.....(((.((((((((((.........)))))))))))))---...))))).)))---........... ( -29.50, z-score = -4.19, R) >droEre2.scaffold_4690 12869620 92 + 18748788 GUCGGC--AUCGAAAGGGCCAAGUGUAUUUUUAGGUGGCAAACAAACGCACAAAAUUUGUUUGCCACUCGCCGCUACACAGCC---AUAUCACGUAU ...(((--.((....)))))..(((((......(((((((((((((.........)))))))))))))......)))))....---........... ( -30.50, z-score = -3.52, R) >consensus GUUGGCU_UUCGAAAGGGCCAAGUGUAUUUUUAGUUGGCAAACAAACGCACAAAAUUUGUUUGCCACU___CGCAGCACAGAC___AUAUCACGUAU ((((((...((....)))))..((((......((.(((((((((((.........))))))))))))).......)))).))).............. (-23.78 = -23.50 + -0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:27:51 2011