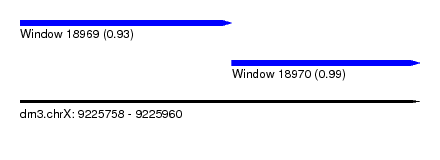
| Sequence ID | dm3.chrX |
|---|---|
| Location | 9,225,758 – 9,225,960 |
| Length | 202 |
| Max. P | 0.991996 |

| Location | 9,225,758 – 9,225,865 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 71.66 |
| Shannon entropy | 0.50895 |
| G+C content | 0.40227 |
| Mean single sequence MFE | -26.11 |
| Consensus MFE | -13.54 |
| Energy contribution | -14.88 |
| Covariance contribution | 1.34 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.934187 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
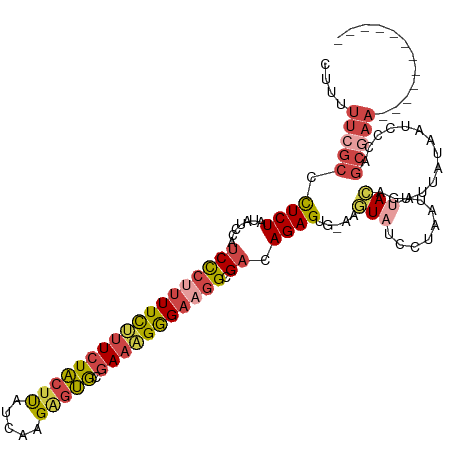
>dm3.chrX 9225758 107 + 22422827 CUUUUUCGCCCUCUAUAUCUAUCCCUUUUCUUUCUACUUUUCACGAGUGCGAAAGGGAAGGCGACAGAGUGGAAGUAUCCAAAUAUACAGAUUACAAUCCCAGCGAA------------ ....(((((..(((((.(((.((((((..((((((((((.....))))).)))))..)))).)).)))))))).((((......))))..............)))))------------ ( -30.60, z-score = -3.29, R) >droAna3.scaffold_13334 1240186 98 - 1562580 UACUUUCAAUUUCUACUGCCAUCUCGCUUUCUUUUGCUUA--AAGGCCACCACGGAGCGAGGGAGAGAGCA-AAAUAAAUUCUUAAAUAAUAUUUAAUCCC------------------ .........................((((((((((((((.--..((...))...))))))..)))))))).-.............................------------------ ( -16.90, z-score = -0.27, R) >droEre2.scaffold_4690 12854840 109 - 18748788 CUUUUUCGCCCUCUAUAUCCAUCUCGUUUCUUUCUACUCAU-AAGAGUGCGAAAGGGAAGGCGCCAGAGGG-AAUUAUCCUAGUUUACAGAUAUUAUUCCCAGCAAAUAGC-------- ......((((................(..((((((((((..-..))))).)))))..).)))).....(((-(((((((..........))))..))))))..........-------- ( -24.03, z-score = -0.63, R) >droYak2.chrX 17818995 118 + 21770863 CUUUUUCGCCCUCUAUAUCCAUCCCUUUUCAUUCUACUUAUCAAGUGUGCGAAGGAGAAGGCGACAGAGUG-AAGUAUCCUAGUUCACAGAUAUUAAUUCCAGCGAAUAUCUAUAUAAC .(((.(((((.(((.........(((((.(((.((........)).))).)))))))).))))).)))(((-((.((...)).)))))(((((((..........)))))))....... ( -24.60, z-score = -1.53, R) >droSec1.super_68 116168 106 + 137979 CUUUUUCGCCCUCUAUAUCUAUCCCUUUUCUUUCUACUUUUCAAGAGUGCGAAAGGGAAGGCGAAAGAGUG-AAGUAUCCUAAUUUACGGAUUAUAAUCCCAGCGAA------------ ....(((((........(((.((((((..((((((((((.....))))).)))))..)))).)).)))(((-((.........)))))((((....))))..)))))------------ ( -30.70, z-score = -2.87, R) >droSim1.chrX 7354770 108 + 17042790 CUUUUUCGCCCUCUAUAGCUAUCCCUUUUCUUUCUACUUUUCAAGAGUGCGAAAGGGAAGGCGACAGAGUG-AAGUAUCCUAAUUUACGGAUUAUAAUCCCAGCGAAUU---------- ....(((((.((((...(((.((((((((..((((........))))...)))))))).)))...))))..-..(((........)))((((....))))..)))))..---------- ( -29.80, z-score = -2.37, R) >consensus CUUUUUCGCCCUCUAUAUCCAUCCCUUUUCUUUCUACUUAUCAAGAGUGCGAAAGGGAAGGCGACAGAGUG_AAGUAUCCUAAUUUACAGAUUAUAAUCCCAGCGAA____________ ....(((((.((((.......((((((((((((((((((.....))))).))))))))))).)).)))).....(((........)))..............)))))............ (-13.54 = -14.88 + 1.34)


| Location | 9,225,865 – 9,225,960 |
|---|---|
| Length | 95 |
| Sequences | 3 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 70.61 |
| Shannon entropy | 0.42186 |
| G+C content | 0.32745 |
| Mean single sequence MFE | -21.52 |
| Consensus MFE | -11.80 |
| Energy contribution | -12.37 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.51 |
| SVM RNA-class probability | 0.991996 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
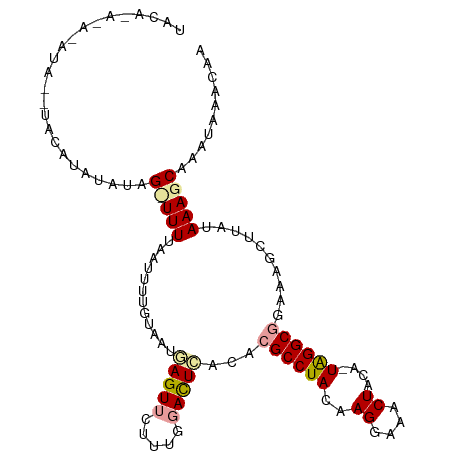
>dm3.chrX 9225865 95 + 22422827 --------------UACACAUAUAG-UUUUAGUUUUAAUAGAAGUCCUUGGAAUUUACAAGCCUACAAGGAAACUACAAUAGGCGAUAAGCCCAUAAAGCAAAUAAACAA --------------..........(-(((..((((((.(((...((((((...............))))))..))).....(((.....)))..))))))....)))).. ( -17.46, z-score = -2.44, R) >droEre2.scaffold_4690 12854949 109 - 18748788 UACAUACAUAUAUGUACAUACUUAGUUUUUAAUUUUGUAAUGAGUUGUUUGGACUGACACGCCUACAAGAAACCUGCA-UAGGCGGAAAGCUUAUAAAGCAAAUAAACAA ((((((....))))))........((((.....(((((.((((((((((......))).((((((..((....))...-))))))...)))))))...))))).)))).. ( -21.50, z-score = -1.41, R) >droYak2.chrX 17819113 110 + 21770863 UACACAUACAUACAUACAUAUAUAGCUUUCAAUUUCGUAAUGAGUUCUUUGGACUCACACGCCUACAAGGAAACUAUAGUGGGCGGCAAACUUAUAAAGCAAAUAAACAA ........................(((((...........((((((.....))))))..(((((((.((....))...)))))))..........))))).......... ( -25.60, z-score = -4.55, R) >consensus UACA_A_A_AUA__UACAUAUAUAG_UUUUAAUUUUGUAAUGAGUUCUUUGGACUCACACGCCUACAAGGAAACUACA_UAGGCGGAAAGCUUAUAAAGCAAAUAAACAA ........................(((((............(((((.....)))))...((((((..((....))....))))))..........))))).......... (-11.80 = -12.37 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:27:49 2011