
| Sequence ID | dm3.chrX |
|---|---|
| Location | 9,208,866 – 9,208,965 |
| Length | 99 |
| Max. P | 0.598516 |

| Location | 9,208,866 – 9,208,965 |
|---|---|
| Length | 99 |
| Sequences | 9 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 67.72 |
| Shannon entropy | 0.63553 |
| G+C content | 0.47775 |
| Mean single sequence MFE | -24.21 |
| Consensus MFE | -8.15 |
| Energy contribution | -7.87 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.598516 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 9208866 99 + 22422827 -----CAACGGUUGC-AGUGUAAAUUGGUCAACUAAAUUAUGCAGCAUCAGUUGCAGUUUGUGCAGUUCCAGUUGCAGCCGAGUCCUUUGUCCUUUGUUCCGCAA-- -----...(((((((-(......(((((..((((..((..((((((....))))))....))..)))))))))))))))))........................-- ( -25.60, z-score = -1.17, R) >droSim1.chrX_random 276925 97 + 5698898 -----CAACGGUUGC-AGUGUAAAUUGGUCAACUAAAUUAUGCAGCAUCAGUUGCAGUUUG--CAGUUCCAGUUGCAGCCGAGUCCUCUGUCCUCCGUUCCGCAA-- -----.(((((..((-((.(....(((((...........((((((....))))))...((--(((......))))))))))...).))))...)))))......-- ( -25.80, z-score = -1.83, R) >droSec1.super_68 99318 97 + 137979 -----CAACGGUUGC-AGUGUAAAUUGGUCAACUAAAUUAUGCAGCAUCAGUUGCAGUUUG--CAGUUCCAGUUGCAGCCGAGUCCUUUGUCCUCCCUUCCGCAA-- -----...(((((((-(......(((((..((((......((((((....)))))).....--.)))))))))))))))))........................-- ( -25.50, z-score = -1.94, R) >dp4.chrXL_group1a 1352267 98 - 9151740 -----AAACGGUUUC-AGUGUAAAUUGGUCAACUAAAUUAUGCAGCAUCAGUUACAGUUC---CAGCGGCAGCAACGGCCAAGCUGUCGAAGCCCUCCUUCCCAGUC -----....((((((-.((((((.((((....)))).)))))).................---....((((((.........))))))))))))............. ( -23.20, z-score = -1.18, R) >droPer1.super_15 2031668 98 - 2181545 -----AAACGGUUUC-AGUGUAAAUUGGUCAACUAAAUUAUGCAGCAUCAGUUACAGUUC---CAGCGACAGCAACGGCCAAGCUGUCGAAGCCCUCCUUCCCAGUC -----....((((((-.((((((.((((....)))).)))))).................---....((((((.........))))))))))))............. ( -23.80, z-score = -1.89, R) >droWil1.scaffold_181096 9547769 99 - 12416693 CAAACGGACGGUUGC-AGUGUAAAUUGGUCAACUAAAUUAUGCAGCAUCAGUUGCAGUU---GCAGCAACAUUCGGGGGCA--UUCCAUUCUCGGCAUUCCACAC-- .....(((..(((((-((.((((.((((....)))).))))(((((....)))))..))---))))).....(((((((..--.....)))))))...)))....-- ( -24.70, z-score = -1.02, R) >droVir3.scaffold_12970 3328689 95 - 11907090 -----AAACGGUUGCCAGUGUAAAUUGGUCAACUAAAUUAUGCAGCAUCAGUUGCACCAG---UAGUCAAGCUGUCAGUUUG----CUCCCGGCCUAGCCUUAGCCC -----....(((((((.(.(((((((((............((((((....)))))).(((---(......))))))))))))----).)..)))..))))....... ( -26.10, z-score = -1.53, R) >droMoj3.scaffold_6482 678801 99 + 2735782 -----AAACGGUUUCCAGUGUAAAUUGGUCAACUAAAUUAUGCAACAUCAGUUGCACCAG---UAGUCAAGCUGUCAGUUCAACCACCCCCGCCGCAGCCCUAGCCC -----....((((..(.(((.....((((.((((......((((((....)))))).(((---(......))))..))))..))))....))).).))))....... ( -19.30, z-score = -1.27, R) >droGri2.scaffold_15203 660150 95 - 11997470 -----AAACGGUUGCCAGUGUAAAUUGGUCAACUAAAUAAUGCAGCAUCAGUUGCACCAG---UAGUCAAGCUGUCAGUUUG----CCACAGAUACCCCUUCCGUCC -----..((((......((((....((((.((((......((((((....)))))).(((---(......))))..)))).)----)))...)))).....)))).. ( -23.90, z-score = -2.30, R) >consensus _____AAACGGUUGC_AGUGUAAAUUGGUCAACUAAAUUAUGCAGCAUCAGUUGCAGUUG___CAGUCACAGUGGCAGCCAAGUCCUCUCAGCCCCCUUCCCCAA_C .........((((......((((.((((....)))).))))(((((....))))).....................))))........................... ( -8.15 = -7.87 + -0.28)

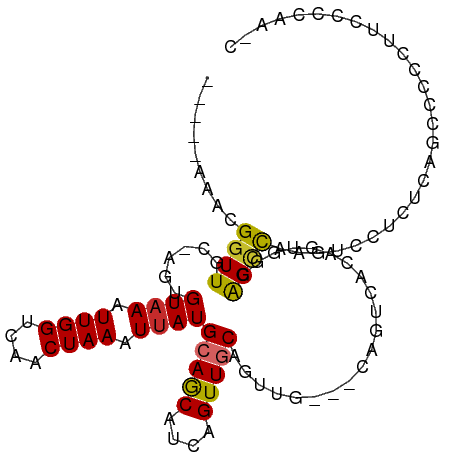

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:27:46 2011