
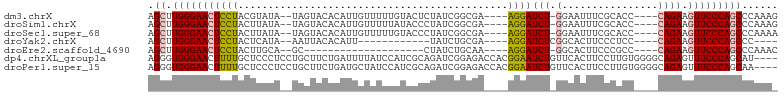
| Sequence ID | dm3.chrX |
|---|---|
| Location | 9,185,172 – 9,185,266 |
| Length | 94 |
| Max. P | 0.993126 |

| Location | 9,185,172 – 9,185,266 |
|---|---|
| Length | 94 |
| Sequences | 7 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 63.44 |
| Shannon entropy | 0.66518 |
| G+C content | 0.49261 |
| Mean single sequence MFE | -28.82 |
| Consensus MFE | -8.94 |
| Energy contribution | -8.49 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.31 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.91 |
| SVM RNA-class probability | 0.974300 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 9185172 94 + 22422827 AGCUUGGGAACUCCUACGUAUA--UAGUACACAUUGUUUUUGUACUCUAUCGGCGA----AGGAUCU-GGAAUUUCGCACC----CAGAAGUUCCCAGCCCAAAG .(((.((((((((((.((((((--.((((((.........)))))).)))..))).----))))(((-((..........)----)))).)))))))))...... ( -31.60, z-score = -3.96, R) >droSim1.chrX 7336627 94 + 17042790 AGCUUGGGAACUCCUACUUAUA--UAGUACACAUUGUUUUUAUACCCUAUCGGCGA----AGGAUCU-GGAAUUUCGCACC----CAGAAGUUCCCAGCCCAAAG .(((.(((((((((((((....--.))).....(((((..((.....))..)))))----))))(((-((..........)----)))).)))))))))...... ( -23.60, z-score = -1.92, R) >droSec1.super_68 77045 94 + 137979 AGCUUGGGAACUCCUACUUAUA--UAGUACACAUUGUUUUUGUACCCUAUCGGCGA----AGGAUCU-GGAAUUUCGCACC----CAGAAGUUCCCAGCCCAAAA .(((.((((((((((.((.(((--..(((((.........)))))..))).))...----))))(((-((..........)----)))).)))))))))...... ( -26.60, z-score = -2.48, R) >droYak2.chrX 17777715 79 + 21770863 AGCUUGGGAACUCCUACUCAUA--AAUUACACAUU------------UAUCUGCGA----AGGAUCUCGGCACUUCCCUCC----CAGAAGUUCCCAGCCC---- .(((.(((((((........((--(((.....)))------------))((((.((----.(((..........))).)).----))))))))))))))..---- ( -21.70, z-score = -2.90, R) >droEre2.scaffold_4690 12814418 74 - 18748788 AGCUUGGGAACUCCUACUUGCA--GC--------------------CUAUCUGCAA----AGGAUCU-GGCACUUCCCGCC----CAGAAGUUCCCAGCCCAAAC .(((.((((((((((..(((((--(.--------------------....))))))----))))(((-(((.......).)----)))).)))))))))...... ( -29.10, z-score = -3.98, R) >dp4.chrXL_group1a 1326478 101 - 9151740 AGGGUGGGAACUUUUGCUCCCUCCUGCUUCUGAUUUUAUCCAUCGCAGAUCGGAGACCACGGAAUCUGUUCACUUCCUUGUGGGGCAGAGUUUCCCAGCAU---- ..(.((((((.(((((((((((((....((((.............))))..))))..((.((((.........)))).)).))))))))).)))))).)..---- ( -33.82, z-score = -1.14, R) >droPer1.super_15 2006185 101 - 2181545 AGGGUGGGAACUUUUGCUCCCUCCUGCUUCUGAUGCUAUCCAUCGCAGAUCGGAGACCACGGAAUCUGUUCACUUCCUUGUGGGGCAGAGUUUCCCAGCAA---- ..(.((((((.(((((((((((((..((..(((((.....))))).))...))))..((.((((.........)))).)).))))))))).)))))).)..---- ( -35.30, z-score = -1.06, R) >consensus AGCUUGGGAACUCCUACUUAUA__UAGUACACAUUGUUUUUAUACCCUAUCGGCGA____AGGAUCU_GGCACUUCCCACC____CAGAAGUUCCCAGCCCAAA_ .((.(((((((((((.............................................)))).........(((...........))))))))))))...... ( -8.94 = -8.49 + -0.44)


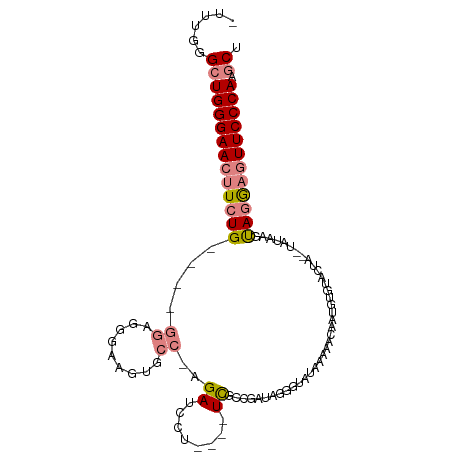
| Location | 9,185,172 – 9,185,266 |
|---|---|
| Length | 94 |
| Sequences | 7 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 63.44 |
| Shannon entropy | 0.66518 |
| G+C content | 0.49261 |
| Mean single sequence MFE | -31.79 |
| Consensus MFE | -12.58 |
| Energy contribution | -13.32 |
| Covariance contribution | 0.74 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.59 |
| SVM RNA-class probability | 0.993126 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 9185172 94 - 22422827 CUUUGGGCUGGGAACUUCUG----GGUGCGAAAUUCC-AGAUCCU----UCGCCGAUAGAGUACAAAAACAAUGUGUACUA--UAUACGUAGGAGUUCCCAAGCU .....(((((((((((((((----.(((((((.....-......)----))))..(((.((((((.........)))))).--))))).))))))))))).)))) ( -33.80, z-score = -3.69, R) >droSim1.chrX 7336627 94 - 17042790 CUUUGGGCUGGGAACUUCUG----GGUGCGAAAUUCC-AGAUCCU----UCGCCGAUAGGGUAUAAAAACAAUGUGUACUA--UAUAAGUAGGAGUUCCCAAGCU .....((((((((((.((((----((........)))-)))((((----.(....((((..((((.......))))..)))--)....).)))))))))).)))) ( -31.20, z-score = -2.58, R) >droSec1.super_68 77045 94 - 137979 UUUUGGGCUGGGAACUUCUG----GGUGCGAAAUUCC-AGAUCCU----UCGCCGAUAGGGUACAAAAACAAUGUGUACUA--UAUAAGUAGGAGUUCCCAAGCU .....((((((((((.((((----((........)))-)))((((----.(....((((..((((.......))))..)))--)....).)))))))))).)))) ( -33.10, z-score = -3.12, R) >droYak2.chrX 17777715 79 - 21770863 ----GGGCUGGGAACUUCUG----GGAGGGAAGUGCCGAGAUCCU----UCGCAGAUA------------AAUGUGUAAUU--UAUGAGUAGGAGUUCCCAAGCU ----.(((((((((((((((----.((((((..........))))----)).)..(((------------(((.....)))--)))....)))))))))).)))) ( -30.40, z-score = -3.23, R) >droEre2.scaffold_4690 12814418 74 + 18748788 GUUUGGGCUGGGAACUUCUG----GGCGGGAAGUGCC-AGAUCCU----UUGCAGAUAG--------------------GC--UGCAAGUAGGAGUUCCCAAGCU .....(((((((((((((((----((((.....))))-......(----((((((....--------------------.)--))))))))))))))))).)))) ( -33.40, z-score = -2.78, R) >dp4.chrXL_group1a 1326478 101 + 9151740 ----AUGCUGGGAAACUCUGCCCCACAAGGAAGUGAACAGAUUCCGUGGUCUCCGAUCUGCGAUGGAUAAAAUCAGAAGCAGGAGGGAGCAAAAGUUCCCACCCU ----.....(((....(((((.((((..((((.........))))))))..(((.(((...))))))...........))))).((((((....)))))).))). ( -29.60, z-score = -0.47, R) >droPer1.super_15 2006185 101 + 2181545 ----UUGCUGGGAAACUCUGCCCCACAAGGAAGUGAACAGAUUCCGUGGUCUCCGAUCUGCGAUGGAUAGCAUCAGAAGCAGGAGGGAGCAAAAGUUCCCACCCU ----.....(((....(((((.((((..((((.........))))))))............((((.....))))....))))).((((((....)))))).))). ( -31.00, z-score = -0.37, R) >consensus _UUUGGGCUGGGAACUUCUG____GGAGGGAAGUGCC_AGAUCCU____UCGCCGAUAGGGUAUAAAAACAAUGUGUACUA__UAUAAGUAGGAGUUCCCAAGCU ......((((((((((((((.....................................................................)))))))))))).)). (-12.58 = -13.32 + 0.74)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:27:43 2011